
Enhancing apple disease resistance using magnetic nanoparticles carrying artificial miRNAs that suppress a tRNA-derived small RNA targeting a disease-resistant gene #research #PlantCommunications cell.com/plant-commun...
21.12.2025 09:39 — 👍 3 🔁 2 💬 0 📌 0

PlantscRNAdb 4.0: Improved marker identification and annotation under a cell type uniformity for plants #resource #MolecularPlant cell.com/molecular-pl...
25.12.2025 08:08 — 👍 4 🔁 2 💬 0 📌 0

Box 1 (shortened, full legend in paper): Key developments in elucidating hierarchical chromatin organizations in plants
Chromosome territories
Each chromosome occupies a distinct territory within the nucleus, as evidenced by strong intra-chromosomal contacts
in a Hi-C contact map.
Large A/B compartments
Each chromosome is partitioned into large A and B compartments. The heterochromatic B compartment is usually located near the nuclear membrane, whereas the euchromatic A compartment tends to be positioned toward the inner regions of the nucleus.
TAD-like domains and compartment domains
Large A and B compartments can be further partitioned into TAD-like domains and/or compartment domains.
Yin et al. (2023) demonstrated that the Arabidopsis genome is partitioned into compartments, each with different histone modifications, including H3K4me3, H3K27me1, and H3K27me3.
🧬 EXPERT VIEW 🧬
In this review, Lee & Seo highlight the hierarchical organization of plant genomes, emphasizing recent advances in understanding local chromatin structures shaped by cohesins and gene borders as fundamental structural units.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
31.12.2025 19:27 — 👍 23 🔁 7 💬 0 📌 0

Box 1 (shortened, full legend in paper): Key developments in elucidating hierarchical chromatin organizations in plants.
Chromosome territories
Each chromosome occupies a distinct territory within the nucleus, as evidenced by strong intra-chromosomal contacts in a Hi-C contact map.
Large A/B compartments
Each chromosome is partitioned into large A and B compartments. The heterochromatic B compartment is usually located near the nuclear membrane, whereas the euchromatic A compartment tends to be positioned toward the inner regions of the nucleus.
TAD-like domains and compartment domains
Large A and B compartments can be further partitioned into TAD-like domains and/or compartment domains.
🧬 SPECIAL ISSUE REVIEW 🧬
In this review, Lee & Seo highlight the hierarchical organization of plant genomes, emphasizing recent advances in understanding local chromatin structures shaped by cohesins and gene borders as fundamental structural units.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
24.12.2025 10:32 — 👍 29 🔁 13 💬 0 📌 0
📢 Online event: International Society of Root Research - Mini Symposium — Wed 28 Jan 2026, 10:00–17:00 UK (12:00–18:30 UTC). Talks + discussion for anyone into root research & related topics.
@rootscientists.bsky.social @rootarchitecture.bsky.social #roots #RootResearch #PlantScience #Rhizosphere
18.12.2025 15:10 — 👍 7 🔁 5 💬 1 📌 0

Get ready: TAIR12 is coming - Phoenix Bioinformatics
More than two years ago, TAIR took the lead in a volunteer-driven project to reannotate the Arabidopsis thaliana genome
It's our birthday! Twenty-five years ago, in December 2000, the Arabidopsis thaliana genome was published and #TAIR was born.
Now, we're preparing for the release of #TAIR12 early in the new year. What a long way we've come!
bit.ly/3KDJciT
#plantbiology #plantscience #genomics #Arabidopsis
18.12.2025 17:32 — 👍 36 🔁 12 💬 0 📌 0

Leandro Quadrana
🎉 Congratulations to Leandro Quadrana (@leandroquadrana), @CNRS researcher at @IPS2ParisSaclay, for obtaining the prestigious #ERCCoG HosTEome!
#ERC #Genomics #Research #AIinBiology #CNRS #UniversitéParisSaclay #PlantScience
ips2.u-psud.fr/en/articles/...
12.12.2025 11:32 — 👍 14 🔁 3 💬 0 📌 0

In vivo RNA structure influences the translation and stability of plant long noncoding RNAs #research #PlantCommunications cell.com/plant-commun...
28.10.2025 09:35 — 👍 4 🔁 3 💬 0 📌 0

A telomere-to-telomere reference genome assembly of tomato cultivar Heinz 1706 #correspondence #PlantCommunications cell.com/plant-commun...
20.11.2025 06:03 — 👍 0 🔁 1 💬 0 📌 1

Summer School 2026
Registrations are open for the Summer School "Saclay Plant Science" (SPS) 2026 on Plant Single Cell at Versailles ! ips2.u-psud.fr/en/articles/... #PlantScience
27.11.2025 13:39 — 👍 1 🔁 1 💬 0 📌 0

Plants establish beneficial interactions with both fungi and bacterial microbes that they attract from the soil rhizosphere by producing signalling molecules under conditions of nutritional deficiency.
(A) arbuscular mycorrhizal (AM) symbiosis, (B) nitrogen-fixing rhizobial nodulation symbiosis, (C) other beneficial plant-fungi interactions, such as the ectomycorrhizal symbiosis (ECM) and the association with Trichoderma or Mucoromycotina ‘fine root endophytes’ (MFRE), (D) interactions with Plant Growth-Promoting Rhizobacteria (PGPR) Figure generated using BioRender (https://biorender.com/).
A review from F. Frugier' team aiming at identifying the common molecular mechanisms associated to the different beneficial plant-microbe symbiotic and endophytic interactions
ips2.u-psud.fr/en/articles/... #PlantScience
27.11.2025 14:30 — 👍 3 🔁 1 💬 0 📌 0

Dr. Bruno Guillotin
Welcome and congratulations to Bruno Guillotin on joining IPS2 as a permanent researcher!
May the spirit of science always be with you and inspire your future discoveries. #PlantScience
ips2.u-psud.fr/en/articles/...
03.11.2025 17:08 — 👍 14 🔁 7 💬 0 📌 0

HeatDDR is funded by the European Union. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union. Neither the European Union nor the granting authority can be held responsible for them
9 international PhD students, 17 partners, 1 goal: coordinated by Université Paris-Saclay, the HeatDDR Doctoral Network is training the next generation of plant scientists to boost crop resilience to heat and climate change 🌱🔥 ips2.u-psud.fr/en/articles/... #PlantScience
14.11.2025 15:34 — 👍 1 🔁 1 💬 0 📌 0
plantiSMASH 2.0: improvements to detection, annotation, and prioritization of plant biosynthetic gene clusters https://www.biorxiv.org/content/10.1101/2025.10.28.683968v1
29.10.2025 05:47 — 👍 5 🔁 5 💬 0 📌 1

From non-coding to chromatin regulators: VIVIpary and the rise of lncRNAs in plant biology #spotlight #MolecularPlant cell.com/molecular-pl...
25.06.2025 16:42 — 👍 2 🔁 1 💬 0 📌 0

Long noncoding RNAs as molecular architects: Shaping plant functions and physiological plasticity #review #MolecularPlant cell.com/molecular-pl...
11.09.2025 06:39 — 👍 3 🔁 1 💬 0 📌 0

PostDoc position in bioinformatics and artificial intelligence. PDF available upon request.
Interested in #lncRNA and #ArtificiaIntelligence?
In the frame of our recently founded French-Korean bilateral project DHARP, we are recruiting a post-doc in bioinformatics and artificial intelligence in our team at
@ips2parissaclay.bsky.social
Application limit: 01/12/2025
22.10.2025 15:27 — 👍 3 🔁 1 💬 0 📌 0
We were very happy about your stay and the nice exchanges. Next time in Chile!
18.10.2025 09:28 — 👍 1 🔁 0 💬 0 📌 0

Model describing the promoting effect on symbiotic nodulation of TDIF signaling peptides through increasing the root stele diameter and adapting the primary metabolism
Have a look to the new study from the Frugier team at IPS2, about the sTDIF signaling peptide that modulates the root stele diameter and primary metabolism to accommodate symbiotic nodulation. ips2.u-psud.fr/en/articles/... #PlantScience
22.08.2025 12:54 — 👍 2 🔁 1 💬 0 📌 0

Out after peer review, collaborative study from Nordborg & Weigel labs with help from many others. Not the largest collection of new Arabidopsis thaliana genomes, but we hopefully put forward some good ideas for how to think about pangenomes and their analysis!
www.nature.com/articles/s41...
20.08.2025 06:23 — 👍 146 🔁 73 💬 1 📌 0
#RNAsky #RNAbiology #PlantScience 🌾🧪
09.08.2025 09:13 — 👍 2 🔁 1 💬 0 📌 0

Please DM me for full text
PhD Position: Root System Architecture and Plant Adaptation to Climate Change at the Institute of Plant Sciences of Montpellier (IPSiM) and the PhenoMEn team at the Amélioration Génétique et Adaptations des Plantes méditerranéennes et tropicales (AGAP)
#PlantSciJobs
06.08.2025 15:10 — 👍 3 🔁 5 💬 1 📌 0
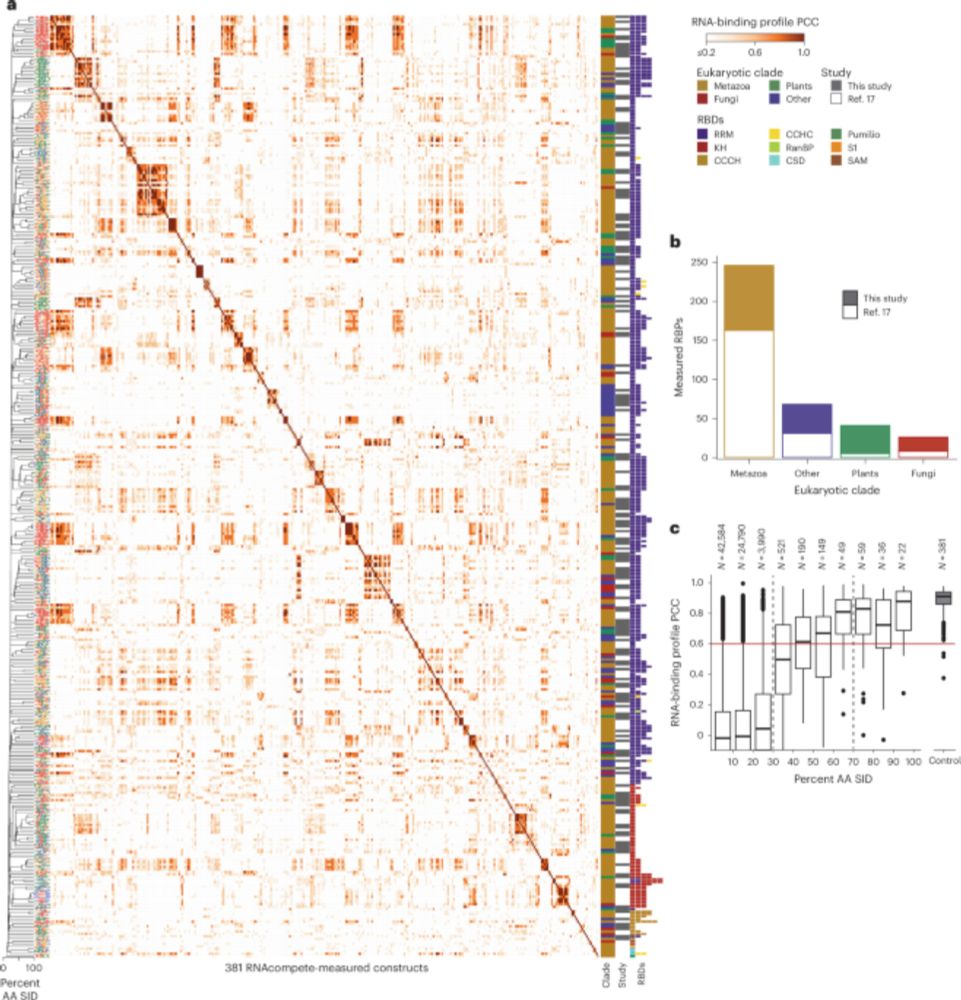
A resource of RNA-binding protein motifs across eukaryotes reveals evolutionary dynamics and gene-regulatory function - Nature Biotechnology
RNA-binding motifs in eukaryotic proteins are presented in a comprehensive resource.
New in @natbiotech.nature.com: a team led by scientists at MSK and University of Toronto have developed a new platform to facilitate the study of RNA-biding proteins (RBPs) across hundreds of species of plants, insects, and animals.
28.07.2025 15:34 — 👍 43 🔁 17 💬 0 📌 3
Nonprofit dedicated to supporting the long-term preservation and usability of research data and other digital research resources. Home of @tairnews.bsky.social and @morphobank.bsky.social
Plant science researcher 🌱 | Root development, cell polarity & cytoskeleton 🌿 | Carbon allocation (source–sink) | Assistant Feature Editor @ Plant Phys | Staying active 🏀🎾, 📚 & ☕️
The Oxford RNA Club is a dynamic community of researchers focused on all aspects of RNA biology, from gene regulation to disease. We host in-person monthly seminars, and larger events throughout the year. Join us here: https://rna.web.ox.ac.uk/home
CNRS group leader & directeur de recherche
Institut de biologie moléculaire des plantes (IBMP)
Strasbourg, France
http://www.ibmp.cnrs.fr/teams/mechanisms-of-small-rna-biogenesis-and-action/?lang=en
We found a strong correlation between effort and disappointment.
https://ko-fi.com/minorrevision <- If you are wondering how these comics are made, it is explained here. No need to have a meltdown.
Tenured scientist @ihsmumacsic.bsky.social Curious about how plants adjust translation to adapt to their environment 👩🏻🔬🍀Science-Dog-📚-Travel-Nature-🌙-lover
The PGRP2026 will take place in the beautiful city of Málaga (Spain)
5 - 7 Oct 2026
Organizers: Laura Arribas-Hernández, Diego López-Márquez, Pablo Manavella, and Catharina Merchante
IHSM-UMA-CSIC
Doctoral student in the Plant Reproductive Biology lab at the Leibniz Institute of Plant Genetics and Crop Plant Research. 🌱
An academic in the School of BioSciences, the University of Melbourne.
The profile photo? Trying to clear out lab fridges late in 2024, and found this enzyme that arrived in January 1992. But don't worry, we haven't thrown it away yet...
gearaguirang.com
Postdoc 📍 Uni Hamburg | When words fail, I speak music | 1st Gen Scientist | she/her 🇵🇭
PhD student @ Weizmann, Plant & Environmental Science Dept.
Biodiversity | Evolution | Psychedelics
COO @ Little Big Science
International Affairs Director @ Survivors of Sexual Assault Advocacy Group
HHMI Hanna Gray Fellow | Asst. Professor @ WashU & Asst. Member @ Danforth Center | Spatial Biology | Plant-microbe Interactions | Duckweed | YouTuber/Streamer | www.thecoxlab.org/
UKRI-Future Leaders Fellow/Group Leader, University of Essex, UK
https://www.thepallavisinghlab.com/
(She/her) 🇮🇳in🇬🇧
#Photosynthesis
#rice #grafting #WaterUseEfficiency #PlantSciences
NIPGR, India - Cornell, USA - Cambridge, UK - Essex, UK
UC Davis rice geneticist & co-author of Tomorrow's Table: Organic Farming Genetics & the Future of Food. TED talk. My lab studies genetics for crop resilience & #plant-microbe interactions. Volunteer Ranger USFS. Nature photography
Plant Ecophysiology Group Leader, Copenhagen Uni. Role & evolution of plant metabolic pathways. Aussie expat, mum of three. Dismantle the tower ✊🏾🌈
One of the longest-running grassland biodiversity experiments worldwide (since 2002).
Uncovering the mechanisms that drive biodiversity-ecosystem functioning (BEF) relationships over time.
https://the-jena-experiment.de
Funding: @dfgpublic.bsky.social
Microscopic farmer 🔬🌺
Microscopy core director @UNC Chapel Hill
Former plant developmental biologist























