
A big boost for cutting-edge research at #McGill 🙌
19 McGill researchers were awarded Canada Research Chairs (new + renewed) supported by a $13M investment from the Government of Canada through the #CRC program 🎉👏 mcgill.ca/x/izt
@waldispuhl.bsky.social
Associate Professor of Computer Science at McGill University. Bioinformatics & Video Games.

A big boost for cutting-edge research at #McGill 🙌
19 McGill researchers were awarded Canada Research Chairs (new + renewed) supported by a $13M investment from the Government of Canada through the #CRC program 🎉👏 mcgill.ca/x/izt
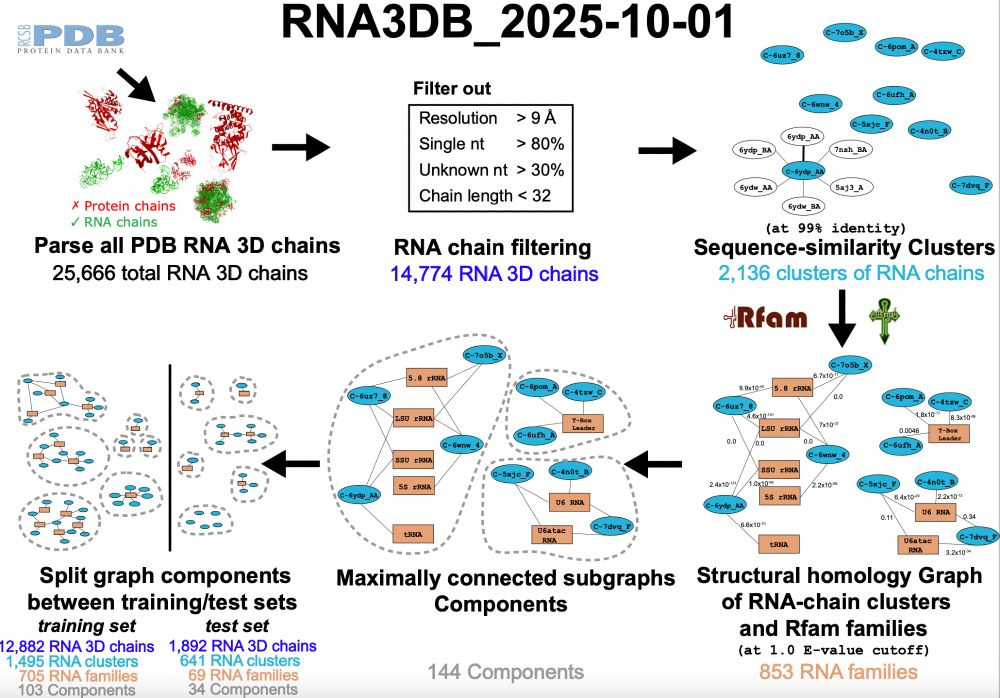
RNA3DB 2025-10-01 release
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).

Integrated prediction of RNA secondary structure jointly with 3D motifs and pseudoknots guided by evolutionary information.
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
Are you looking for a lectureship in biochemistry, molecular genetics, or bioinformatics? Come and enjoy the greatest place for kayak fishing and a fun research+teaching career.
*there are other fun things to do here too.

@d2rmcgill.bsky.social will host a seminar of Eric Westhof on Sep 24. Join us if you want to discuss where we stand in RNA structure prediction!
www.mcgill.ca/dna-to-rna/c...
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...

www.nature.com/articles/s41... From remote matches to @rfamdb.bsky.social models to the discovery of novel *vertebrate* riboswitches!
19.08.2025 01:20 — 👍 5 🔁 3 💬 1 📌 0
McGill’s Joelle Pineau on the work she did as the VP of AI research at Meta, why Canada needs to plan for its digital sovereignty, and why she has little patience for “sensational claims” about how “we’re in imminent peril of a technological apocalypse” universityaffairs.ca/features/for...
23.07.2025 18:44 — 👍 4 🔁 1 💬 0 📌 0
🚨 @healthsciences.mcgill.ca is recruiting: Canada Excellence Research Chair in #RNA Neurotherapeutics
Looking for an established leader in RNA research in the brain with an innovative research program in RNA-related research in neuropsychiatry.
✍ Apply by July 4:




Génome Québec est heureux de lancer le 5e cycle du Programme d’intégration de la #génomique – #agriculture et #bioalimentaire, #foresterie et #environnement.
La date limite pour soumettre une demande est le 20 octobre 2025. Pour plus d’information 👉 rb.gy/kjs0vq

Are you a McGill student interested in developing your science communications skills? We’re looking for a communications intern to help us build awareness of Faculty of Science research, students, publications, events, and other initiatives.
Apply by June 19 ➡️ mcgill.ca/x/iQK
Phage people: Rich Losick and I are combing the world looking for T4 rIIB mutant FC0 (also known as P13). FC0 was the starting point for Francis Crick's beautiful 1961 paper on the triplet nature of the genetic code. We want to sequence it. Anyone have it in an ancient stock collection?
01.06.2025 14:46 — 👍 63 🔁 79 💬 4 📌 5🧬🌊 code Béluga : un projet de science participative pour mieux comprendre le fleuve Saint-Laurent.
Participez aux collectes d’eau de mai à août 2025!
🎥 Pour regarder notre capsule vidéo 👉 rb.gy/sakf14
📍 Infos et lieux d’échantillonnage 👉 rb.gy/vyk0n9

Our first panel talk is LIVE!
Jérôme Waldispühl kicks things off with Leveling Up Citizen Science for (meta)genomic research
@waldispuhl.bsky.social
Join #MVIF 38 👇
🔗 cassyni.com/s/mvif-38
#Microbiome #MeetTheSpeakers

MVIF 38
NEXT WEEK don't miss #MVIF 38 on Microbiome and Citizen Science
👉https://cassyni.com/s/mvif-38
& meet:
🇫🇷 @patrickveiga.bsky.social & Mathieu Almeida
🇮🇹Claudia Cappello
🇨🇦 @waldispuhl.bsky.social
🇺🇸Daniel McDonald
🇺🇸Mikayla A. Borton
🇧🇪Sarah Ahannach
🇳🇱Saara Suominen
🇺🇸 @gilbertjacka.bsky.social

MVIF 38
It’s Friday
...and a new #MVIF program is out! 🤩
Register👉 cassyni.com/s/mvif-38
Contributed talks by
🇫🇷 @patrickveiga.bsky.social
🇮🇹 Claudia Cappello
Keynotes:
🇨🇦 @waldispuhl.bsky.social
🇺🇸Daniel McDonald
🇺🇸Mikayla A. Borton
🇧🇪Sarah Ahannach
Moderators:
🇳🇱 Saara Suominen
🇺🇸 @gilbertjacka.bsky.social

Our latest work in RNA bioinformatics was published today: www.nature.com/articles/s41...
If you are looking for a tool to speed up RNA virtual screening, this is for you.
Congrats to the team and thank you to @genomequebec.bsky.social , FRQNT, and NSERC for the support!
@mcgillscience.bsky.social

Visit the Faculty of Science crowdfunding page to learn about some of the great student-run initiatives raising money this #McGill24, like the McGill Biochemistry Undergraduate Society (BUGS)!
➡️ crowdfunding.mcgill.ca/ui/main/t/sc...

Another really interesting microbiome paper worth checking out www.cell.com/cell/fulltex...
22.01.2025 17:44 — 👍 37 🔁 12 💬 2 📌 0
The January issue is live nature.com/nbt/volumes/...
On our cover, a minigame integrated within a popular video game let more than 4 million players contribute to improving a multiple sequence alignment for microbial phylogeny estimations and association go.nature.com/4aSfOg6

Tertiary structure comparisons between 23 experimentally determined small RNA structures and the best structure prediction selected from 18 different groups.
The latest RNA-Puzzles paper is out (CASP for RNA structure prediction). Check how wild some of the best predictions are for these small RNAs. We're still a long way from a reliable AlphaFold for RNA.
www.nature.com/articles/s41...