Hope you and your family have a lovely day, looks beautiful!
13.10.2025 15:15 — 👍 1 🔁 0 💬 0 📌 0
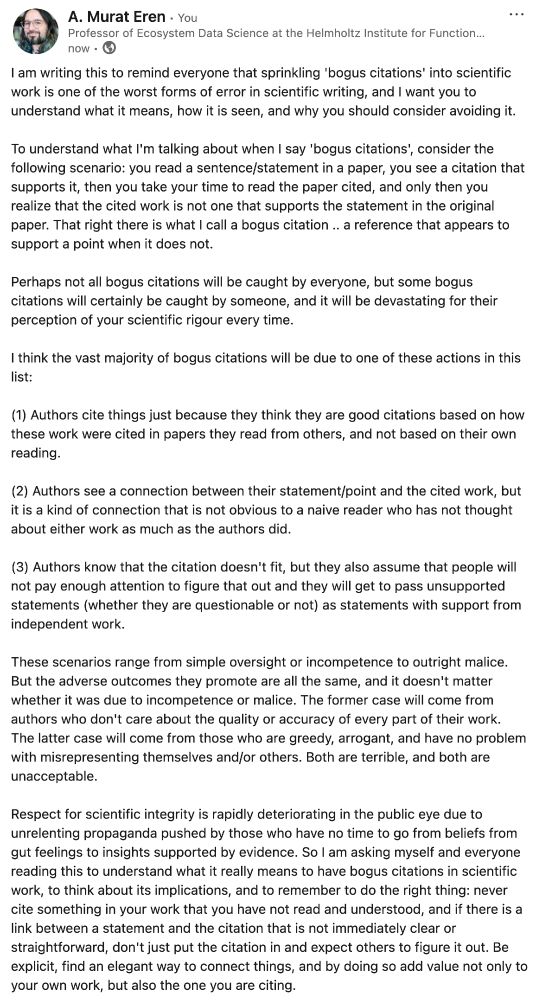
Not that you care, but I still wanted to let you know that if I see a paper full of bogus citations with your name on it, I'm not talking to you anymore.
I was so triggered by a paper I was reading this morning, I am drinking my second coffee.
30.05.2025 09:03 — 👍 33 🔁 7 💬 4 📌 4
Human reference microbiome profiles of different body habitats in healthy individuals
Oh and Park (2025): Human reference microbiome profiles of different body habitats in healthy individuals.
…the differences they identify using logFC between different body sites can be used clinically to identify the health status of an individual. We felt that the study needed a much larger sample size before this would be appropriate.
You can read our full discussion here: dalmug.org/blood-refere...
17.04.2025 18:00 — 👍 0 🔁 0 💬 0 📌 0
We found the alpha diversity results to be particularly striking: they find that alpha diversity follows the pattern blood > saliva > stool. As this appears to be in contrast to most existing literature, we’d have loved to see more discussion of these results.
The authors also suggest that…
17.04.2025 18:00 — 👍 0 🔁 0 💬 1 📌 0
Frontiers | Human reference microbiome profiles of different body habitats in healthy individuals
Last week, we read “Human reference microbiome profiles of different body habitats in healthy individuals” (published here: www.frontiersin.org/journals/cel...)
In this study, the authors characterised the microbiome in blood, saliva and stool of 171 healthy/10 periodontal disease participants.
17.04.2025 18:00 — 👍 0 🔁 0 💬 1 📌 0
DalMUG – Dalhousie Microbiome User Group (DalMUG).
Dalhousie Microbiome User Group (DalMUG).
After a long hiatus, we’re back to writing up blog posts of our DalMUG journal club discussions! You can see all of our previous posts here: dalmug.org
17.04.2025 18:00 — 👍 0 🔁 0 💬 1 📌 0
We also changed the name of the new database - what seemed like a lighthearted joke a few months ago didn’t feel so lighthearted anymore. But the genomes and annotations within the database are not changed, so all of the information above in the thread is still correct.
10.04.2025 12:43 — 👍 0 🔁 0 💬 0 📌 0
I’ve also overhauled the Wiki pages a bit, so I hope it’s easier for users to find the information they need. It’s already available through conda but I’m still working on the Q2 plugin!
10.04.2025 12:43 — 👍 0 🔁 0 💬 1 📌 0
Congratulations!! 🥳
23.02.2025 18:22 — 👍 0 🔁 0 💬 0 📌 0
I’ll be working to get it integrated with the @qiime2.org plugin and available through bioconda soon!
20.02.2025 13:37 — 👍 0 🔁 0 💬 1 📌 0
We’ve also expanded the number of EC number and KO annotations by about 1.3-fold, and it’ll be easy for users to add their own functions to the database with HMMs now.
The new database is included from PICRUSt2 v2.6.0, and if you use it, we’d love to hear your feedback on it!
20.02.2025 13:37 — 👍 0 🔁 0 💬 1 📌 0
This has expanded the number of genomes present from 19,493 to 26,868 bacterial and 406 to 1,002 archaeal. This means that the genomes more closely match study sequences and therefore better represent a range of different environments.
20.02.2025 13:37 — 👍 0 🔁 0 💬 1 📌 0
We now use genomes annotated with Eggnog and phylogenetic trees from GTDB, so we can easily update the genomes as well as the functions included in the future.
20.02.2025 13:37 — 👍 0 🔁 0 💬 1 📌 0
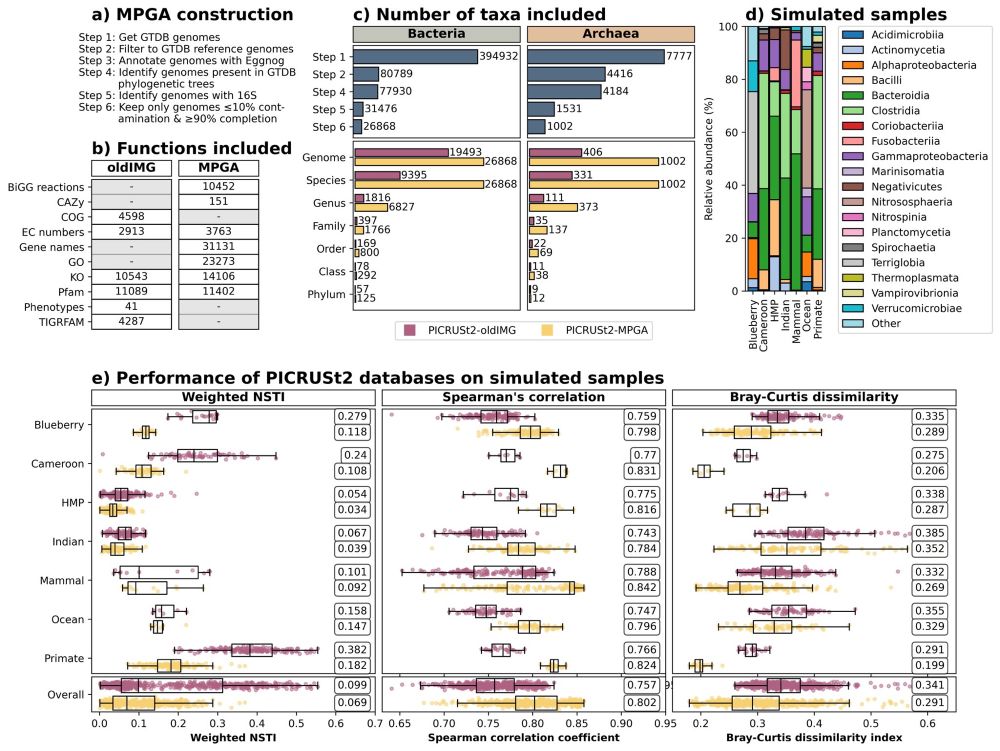
Figure giving an overview of the PICRUSt2 database - Figure 1 from the preprint. It includes the steps taken for database construction, functions included in the previous oldIMG and new MPGA databases, number of taxa included in both databases, composition of simulated samples, and the performance of the oldIMG and MPGA databases on the simulated samples from different environments.
I started thinking about how we could make the PICRUSt2 database easy to update and expand about 5 years ago now, so it’s exciting to finally have something that others can use!
20.02.2025 13:37 — 👍 0 🔁 0 💬 1 📌 0

Big news - part 1 of the 2025 Canadian Bioinformatics Workshop Series has arrived! Learn more and apply here: bioinformatics.ca/workshops/cu...
07.02.2025 22:01 — 👍 5 🔁 2 💬 0 📌 1
Environmental microbiologist, biogeochemical cycles, AMR in the environment
Royal Society Dorothy Hodgkin Research Fellow at UEA
Editor-In-Chief LAM Journal
🏳️🌈ally | mum of 2 | I love 🌋
www.marcelahernandez.com
Microbial ecologist at UniMelb collaborating with conservation scientists, immunologists, and nutritionists to improve conservation outcomes for threatened species.
🐨 Native Australian mammals
🦠 Microbes
🧬 Bioinformatics
Associate Professor of Micobiomes at the University of Maine; Founder of the Microbes and Social Equity working group; Senior Editor at mSystems; Early Career At-Large Member of the Board of the American Society for Microbiology.
Systems Microbial Ecologist | Design Thinker | F1 & IndyCar fan. Metalhead. Studying microbes in Desert Soil and nearshore Marine Sediments.
Blog: uncultured.carinilab.com
Web: CariniLab.com
Linkedin: https://www.linkedin.com/in/paul-carini/
Microbiologist | Director of Genomics Operations @SeqCoast.bsky.social | #bluesoup is best soup | mother to two bundles of chaos | she/her | opinions my own
Algae enthusiast, Associate Professor @UniofExeter, joint appointment @thembauk
Algal ecophysiology | signalling | microbiome | molecular microbiology | diatoms!
Polar & Climate Science / Microbial Ecology / Bioinformatics
Views are my own
He/Him
🏳️🌈🏳️⚧️ ally
christrivedi.com
Science integrity consultant and crowdfunded volunteer, PhD.
Ex-Stanford University. Maddox Prize/Einstein F Award winner
NL/USA/SFO.
#ImageForensics
@MicrobiomDigest on X.
Blog: ScienceIntegrityDigest.com
Support me: https://www.patreon.com/elisabethbik
The image that I run from
Only seems to follow me
Microbiome Scientist, UAMS.
#bioinformatics #microbiome #nutrition #cancer #primaryciliarydyskinesia #rotifers #meiofauna #microfauna Posts are my own. Reposts/likes not necessarily endorsement. 😷
Papa to 9 microbes, mackerel snapper, shepherd, master of sucking. I want to help you learn the skills you'll need to succeed in science.
Postdoc at Dana-Farber and Harvard Med with Heng Li (@lh3lh3.bsky.social). Prev: UBC / UofT.
I like thinking about computational biological sequence analysis and its applications to metagenomics.
https://jim-shaw-bluenote.github.io
Bioinformatician at the Centre for Pathogen Genomics at the University of Melbourne
Professor (Bioinformatics of Small and/or Fishy Things) in Computer Science at Dalhousie University; environmental 🧬 enthusiast. Trying and failing to not post about politics. he/him
The pan-Canadian microbiome research core funded by CIHR. We aim to harness the power of the microbiome to promote human health. https://www.impactt-microbiome.ca/
RNA virus discovery 🦠🔎
Irish Research Council postdoc 👩🏼🔬 at University College Dublin (UCD) 🍀
Ballet and book lover 🩰 | Eurovision fanatic
🇮🇪 🇮🇹 🇩🇰 🇩🇪
Microbiology Society: A world in which the science of #microbiology provides maximum benefit to society | microbiologysociety.org
lnk.bio/microbiosoc
Inupiaq, scientist of host-microbe interactions, mom, runner, news junkie, not necessarily in that order. I speak for myself. She/her. Only reskeets posts with alt text. Unapologetically typo prone.
Microbes & mucus 🤩 | Gut Microbial Ecology | Wageningen University, NL