As mentioned above this is still very early R&D work, not ready for release yet.
We are generating more data and @stoibs11.bsky.social's team has got plenty of ML magic tricks to significantly improve the performance of the models.
28.11.2024 12:11 — 👍 3 🔁 1 💬 1 📌 0
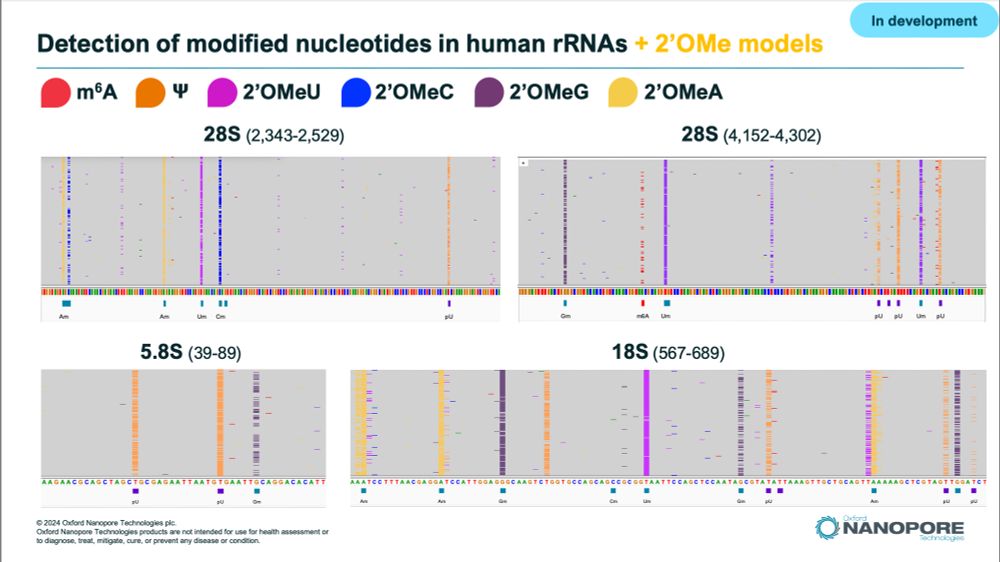
Selected areas of Human rRNAs showing modification calls in IGV. Samples sequenced by ONT direct RNA sequencing with experimental Remora models calling m6A, m5C, PseudoU, and all 4 2'Ome modifications.
We have been busing working on models to detect all 2'Ome-RNA modified nucleotides on top of PseudoU, m6A, m5C and Inosine using @nanoporetech.com direct RNA sequencing.
This is still very preliminary but here are a few examples of what it looks like on Human #rRNA prepared with standard lib prep 🤩
28.11.2024 12:11 — 👍 32 🔁 12 💬 3 📌 0

We are proud to announce a collaboration with UK Biobank to create the world’s first large-scale #epigenetic dataset of 50k participants. The dataset will unlock crucial insights into how #epigenetics drives disease & the breakthroughs to treat them.
Read more: nanoporetech.com/news/oxford-...
27.11.2024 09:16 — 👍 128 🔁 39 💬 1 📌 8
This is worth looking at. Trying some genomes we have assembled with earlier versions of hifiasm, hifiasm plus herro and then this new version of hifiasm. The early assembly results suggest that now hifiasm alone is equivalent to what we got with herro but with less compute!
28.11.2024 12:58 — 👍 21 🔁 5 💬 0 📌 0
Thank you @theoreticalfun.bsky.social and @danrdanny.bsky.social!
14.09.2023 13:29 — 👍 3 🔁 0 💬 0 📌 0
Associate director of modified base machine learning @nanopore | dad of 3 | amateur farmer | Nationals fan | views are my own
Director of Modified Bases Research at @nanoporetech.com
EMBL-EBI alumni, keen cyclist and father of two.
Views are my own. Reposts are not endorsement.
🌍Direct DNA/RNA analysis for anyone, anywhere
🧬Short to ultra-long reads in real-time for rapid insights
🔬Products for research use only
Senior Scientist BCCDC in Microbial Genomics and itinerant Nanopore tinkerer
Public health (meta)genomics & bioinformatics, Prof @unibirmingham.bsky.social, Director @imibirmingham.bsky.social & @climb.ac.uk
UKRI Future Leaders Fellow, University of Birmingham
Finished a human genome, working on a few more 👨💻
Lab: https://genomeinformatics.github.io
Posts are my own
PhD candidate at UCSC BMEB in the Daniel Kim lab interested in transposable elements, metagenomics, and long reads
Sequencing, adapting and exploring genomes.
https://linktr.ee/matt.loose
Enrolled Kawaika (Laguna Pueblo), Haaku (Acoma Pueblo) & Diné (Navajo). Likes to eat, bike, run, & science (chemistry PhD, microbiome, acidophiles). Associate professor of Chemistry at Fort Lewis College; She/hers
CODA. Dad. Interested in genetics, long-read sequencing, and ultramarathon running. Assistant Professor at University of Washington. The command line is my happy place. https://millerlaboratory.com


