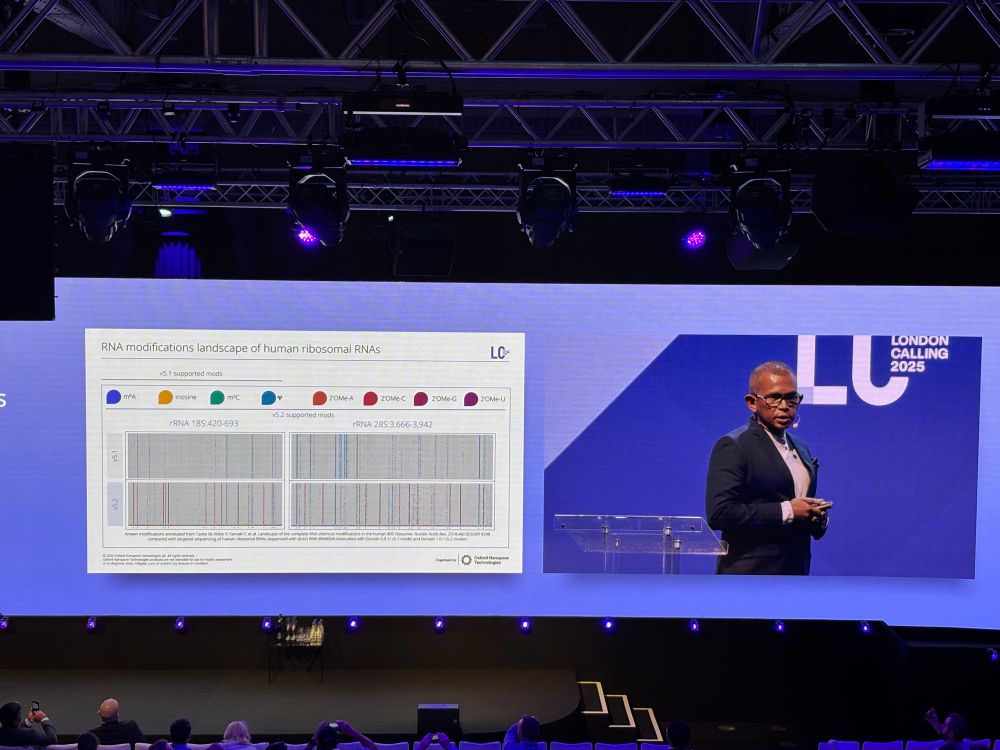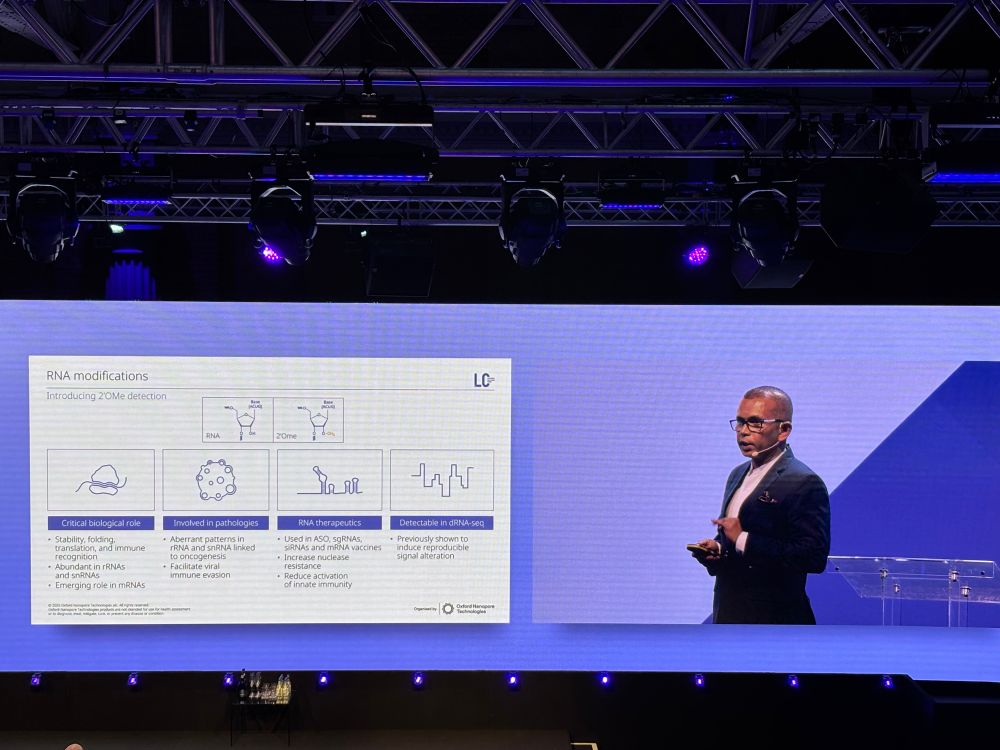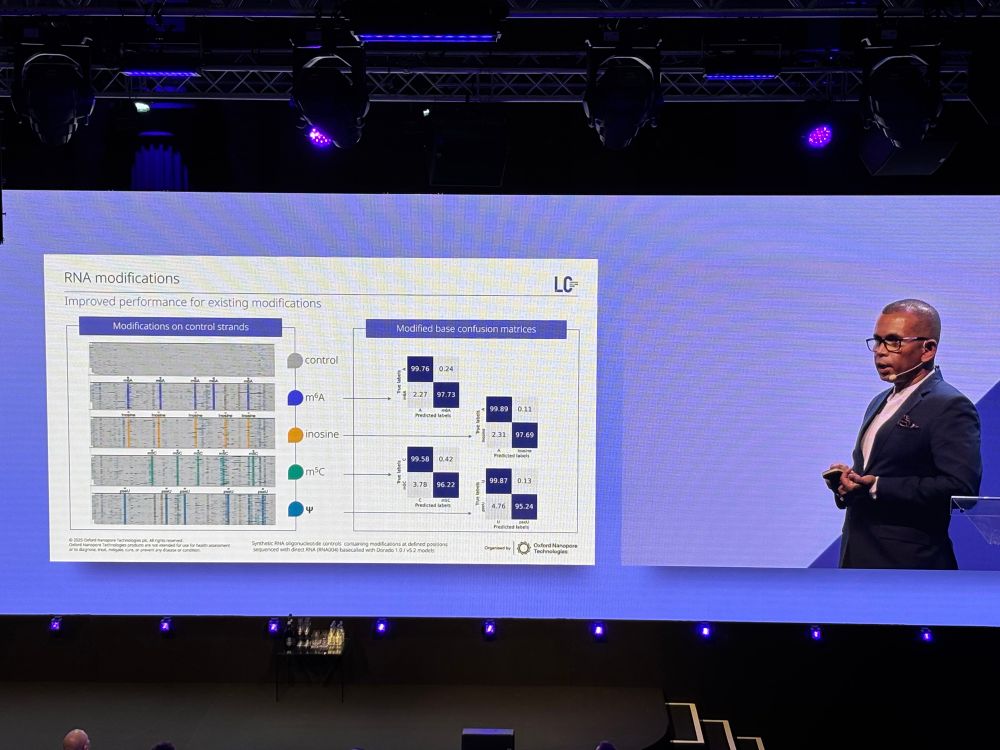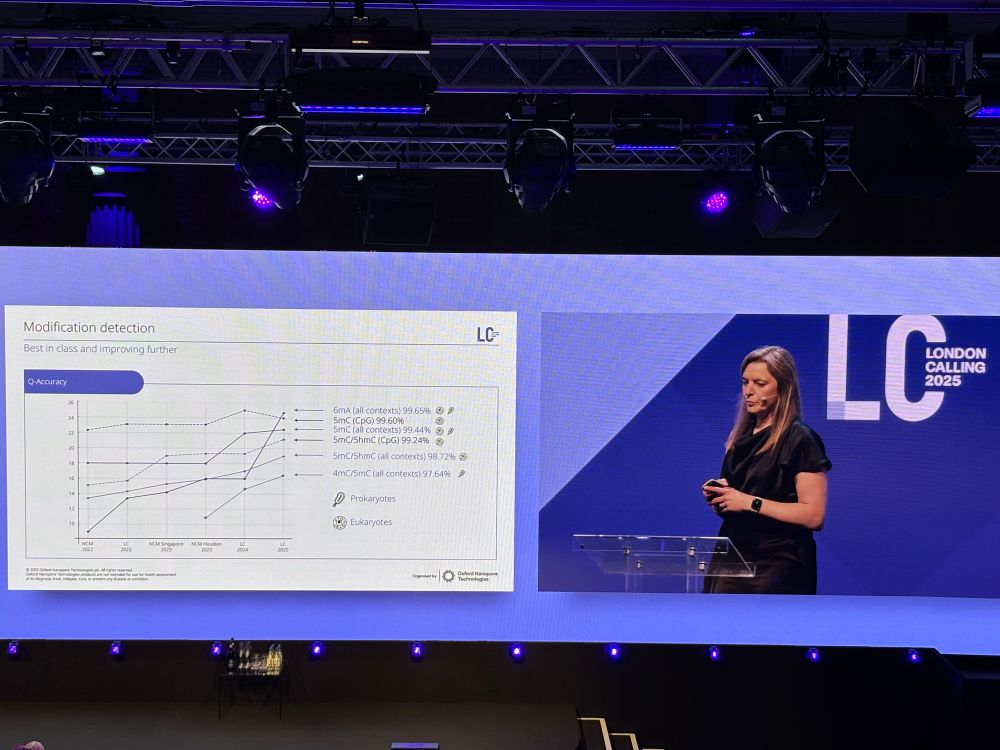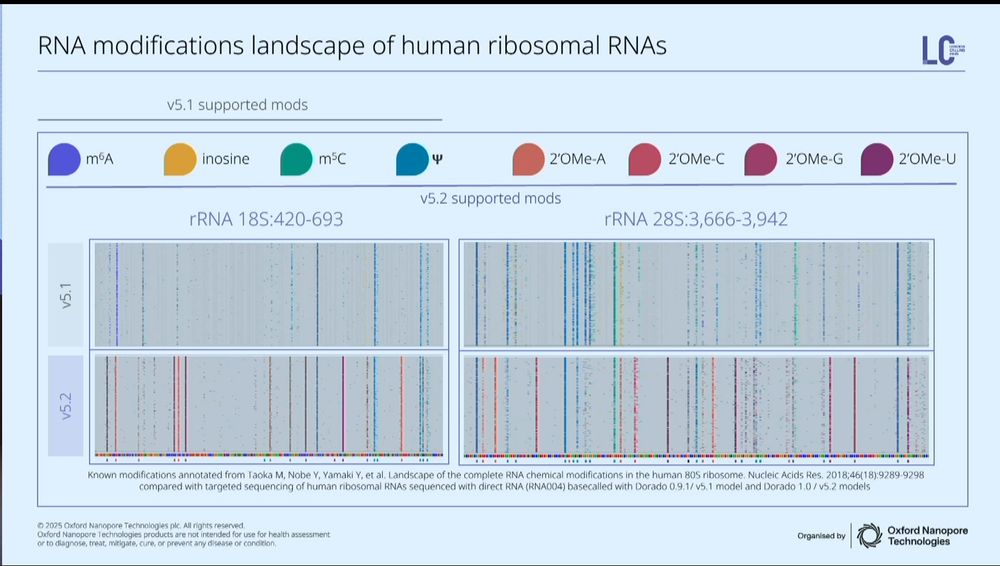
image showing RNA modification landscape of human ribosomal RNAs for 18S and 28S rRNA; patterns of different modifications are shown throughout the entirety of the sequence. A validation of synthetic constructs demonstrates RNA modification detection accuracies of 95-99%.
#NanoporeConf Lakmal Jayasinghe
ONT is the *only* sequencing technology that detects RNA modifications directly. They're now up to 8 modifications that can be detected simultaneously.
[This is something I think ONT should be shouting from the rooftops every day]
RNA barcoding is in beta testing.
22.05.2025 09:57 — 👍 24 🔁 9 💬 1 📌 1

Oxford Nanopore sequencing will be ubiquitous for multiomics….and the next step is proteomics.
First panel-based proteomics assays, before driving towards full protein sensing in the future. #nanoporeconf
21.05.2025 18:27 — 👍 29 🔁 14 💬 2 📌 2


Amazing time to be working with @nanoporetech.com sequencing! The pace of innovation in this network of researchers is astonishing! #NanoporeConf
21.05.2025 17:55 — 👍 3 🔁 2 💬 0 📌 0

Initial release of Pomfret for methylation assisted phase block joining for more contiguous phased assemblies without any additional library prep from @nanoporetech.com data. Such amazing work from Xiaowen Feng! #NanoporeConf
github.com/nanoporetech...
21.05.2025 17:51 — 👍 1 🔁 0 💬 0 📌 0

Exciting modkit release with open chromatin prediction support and more! Amazing work from Art Rand! @nanoporetech.com #NanoporeConf
github.com/nanoporetech...
21.05.2025 17:50 — 👍 2 🔁 0 💬 0 📌 0


Methylation assists tandem repeat expansion estimation from @nanoporetech.com sequencing using strcount. #NanoporeConf
21.05.2025 15:16 — 👍 6 🔁 3 💬 0 📌 0
Amazing work! Congratulations on the publication!
21.05.2025 09:37 — 👍 0 🔁 0 💬 0 📌 0

Excited for another London Calling! Incredible updates coming over the next two days! #NanoporeConf @nanoporetech.com
21.05.2025 08:59 — 👍 7 🔁 1 💬 1 📌 0
Easton about to come out with the Hype “Fire in the Hole” - Torpedo edition!
02.04.2025 03:37 — 👍 0 🔁 0 💬 0 📌 0
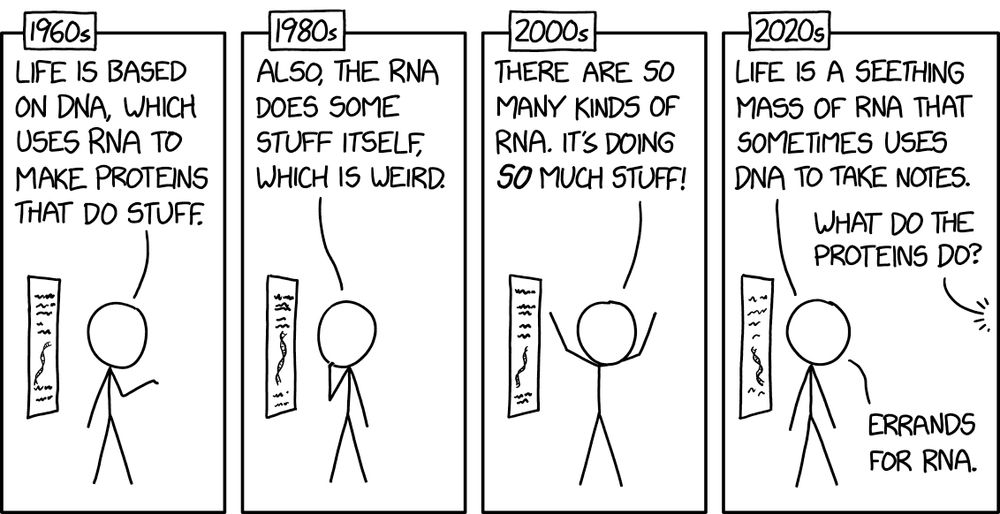
RNA xkcd.com/3056
26.02.2025 14:58 — 👍 17914 🔁 2570 💬 155 📌 171


An overview of nanopore sequencing for pangenomes and hemi-hydroxymethylation in medaka fish brains by @ewanbirney.bsky.social
Presenting results showing biased 5hmC/5mC hemimethylation at splice sites, and increased skewed hemimethylation across repetitive sequences.
23.02.2025 23:08 — 👍 14 🔁 4 💬 1 📌 0
Since Oxford Nanopore sequencing does not require any chemical (bisulfite) conversion to detect methylation. It would mean long-range epigenetic modifications, structural variants (SVs),single nucleotide polymorphisms (SNPs), and repeats can all be identified and phased in a single dataset
27.11.2024 11:10 — 👍 9 🔁 4 💬 0 📌 0

We are proud to announce a collaboration with UK Biobank to create the world’s first large-scale #epigenetic dataset of 50k participants. The dataset will unlock crucial insights into how #epigenetics drives disease & the breakthroughs to treat them.
Read more: nanoporetech.com/news/oxford-...
27.11.2024 09:16 — 👍 128 🔁 39 💬 1 📌 8
Thank u for this question. Worth another thread as it reflects common confusion that more data & better AI/ML is the solution (newer official @nanopore models & new customer methods are being developed/released). For abundant mods like 5mCpG in human, this strategy works. *BUT* for low abundance 1/5
21.11.2024 12:15 — 👍 6 🔁 1 💬 1 📌 0
Table of Contents — November 2024, 34 (11)
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
Long-read special issue of GR is out, including our paper on T2T assembly using only Nanopore. Just in time for ONT to discontinue duplex cells on Nov 27 😅😂 Good thing Verkko also works great with HERRO-corrected simplex data!
📄 genome.cshlp.org/content/34/1...
📖 genome.cshlp.org/content/34/1...
21.11.2024 00:50 — 👍 86 🔁 34 💬 4 📌 0
Journalist and author. My newsletter, the Long Game, is about baseball, creativity, and mental health. You can find it here: https://mollyknight.substack.com. My dog is named Canelo.
Associate Professor
DFCI & HMS
Constitutional law prof and historical political scientist studying:
The United States Supreme Court
American Political Development
Anglo-American Constitutionalism
📍Atlanta
Author, Rot and Revival:
https://www.ucpress.edu/books/rot-and-revival/paper
Husband, dad, veteran, writer, and proud Midwesterner. 19th US Secretary of Transportation and former Mayor of South Bend.
👩🔬 The Institut Pasteur is a leading global biomedical research institute.
🔬 Explore with us the frontiers of biomedical research
🧪 Follow us to uncover groundbreaking discoveries
🌍 Join a community passionate about progress and open science
Get every sports story that matters.
The official BlueSky account of Baseball Prospectus. | Subscribe today for less than $3.83 per month: https://www.baseballprospectus.com/subscriptions/
programming and exclamation marks
blog: jvns.ca
zines: wizardzines.com
Epigenetic Inheritance, Neuroscience & anything biology-related
https://www.odedrechavilab.com/
https://www.qedscience.com
TED: https://shorturl.at/myFTY
Huberman Lab Podcast: https://youtu.be/CDUetQMKM6g
Consensus fantasy football advice, tools and accuracy tracking from 200+ experts to help you dominate 💪
IG & Twitter/X: @FantasyPros
YouTube: http://fntsy.pro/YouTube
CSO at Enhanced Genomics #biotech #biosky #punting
Associate Professor of CS @ University of Maryland. Proud Rust advocate! I ♥ science & compiled, statically-typed programming languages! Views are my own. Tech stack: https://github.com/rob-p/tech-stack.
henfluencer - geriatric millennial - #stopasianhate - don’t call me Clem - emotional eater™️- he/him- @chowlab
https://linktr.ee/clementchow
Assistant prof at UCLA using the Bayes for the genomes https://pimentellab.com
Senior Bioinformatics Scientist @ Oxford Nanopore Technologies ;
Algorithms, complexity, cancer ;
Opinions are my own
Dad. Scientist at VIB. Bioinformatics postdoc. Sequencing technologies enthusiastic. Geneticist. He/him.