
"What James Watson got wrong about DNA"
By the great Sohini Ramachandran (@sramach.bsky.social) and your boy for The Boston Globe (@bostonglobe.com).
www.bostonglobe.com/2025/11/14/o...
@arbelharpak.bsky.social
Evolutionary, statistical and population genetics at UT Austin. http://www.harpaklab.com

"What James Watson got wrong about DNA"
By the great Sohini Ramachandran (@sramach.bsky.social) and your boy for The Boston Globe (@bostonglobe.com).
www.bostonglobe.com/2025/11/14/o...
My department at UT Austin is looking to hire an Assistant Professor in Evolutionary Biology, broadly defined. Feel free to reach out to me with any questions you may have.
apply.interfolio.com/177547
An empirical approach to evaluating the prevalence of long-lived balancing selection in humans--and important limitations. Work by @hannahmm.bsky.social
11.11.2025 19:14 — 👍 62 🔁 35 💬 0 📌 0@jbenning.bsky.social @jedidiahcarlson.com
et al. exemplify how confounding can lead to flawed inference on genetic causality. In studies of human behavior and social outcomes, the cost of downplaying this problem can be steep. royalsocietypublishing.org/doi/epdf/10.... bsky.app/profile/jben...

Why do complex traits differ in their genetic architecture?
In our new PLOS Biology paper, we will try to convince you that two simple scaling laws drive differences in the number, effect sizes and frequencies of causal variants affecting complex traits.
Thread:
journals.plos.org/plosbiology/...

If you are at #ASHG2025 and would like to chat in person, please reach out! My lab is looking for curious scientists to join us, especially (but not limited to) postdoctoral researchers.
14.10.2025 15:45 — 👍 5 🔁 4 💬 0 📌 0Not necessarily a bad thing: perhaps your subscribers could use a little more boring in their lives.
10.10.2025 18:53 — 👍 0 🔁 0 💬 1 📌 0
Delighted that our paper about the distribution of genomic spans of clades/edges in genealogies (ARGs), and using this for detecting inversions and other SVs (and other phenomena that cause local disruption of recombination) is out in MBE academic.oup.com/mbe/article/... (1/n)
03.10.2025 09:54 — 👍 64 🔁 37 💬 4 📌 1Bittersweet to be leaving @docedge.bsky.social after a wonderful postdoc, but excited to share that I'm joining @uoregon.bsky.social next month as an Assistant Professor in the Department of Data Science.
06.08.2025 17:18 — 👍 146 🔁 28 💬 21 📌 4
1/13 In a new preprint, we (with @xliaoyi.bsky.social in the @arbelharpak.bsky.social lab) find that expression from the “inactivated” X (Xi) is consequential for both female-male differences in gene expression and variation among females in disease and physiology: www.biorxiv.org/content/10.1...
06.08.2025 02:42 — 👍 21 🔁 16 💬 2 📌 0
Applications for the 2026 Stengl-Wyer Scholars Program are open! The program provides up to 3 years of support for talented postdoctoral researchers in the broad area of the diversity of life and/or organisms in their natural environments. Learn more here: utexas.infoready4.com/CompetitionS...
05.08.2025 21:09 — 👍 9 🔁 13 💬 0 📌 2Amidst all the terrible and terrifying news, so lovely to hear of
@jkpritch.bsky.social's election to the National Academy of Sciences. Congratulations!
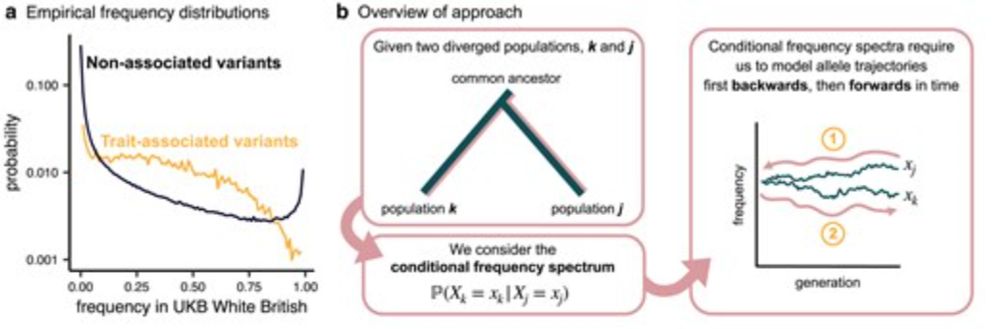
a quick note on my paper with @jeffspence.bsky.social and @jkpritch.bsky.social on conditional frequency spectra, now out in a @genetics-gsa.bsky.social special issue: doi.org/10.1093/genetics/iyae210
18.04.2025 19:49 — 👍 60 🔁 24 💬 3 📌 1
More recently, Smith et al. (biorxiv.org/content/10.1...) proposed an estimator of polygenic score confounding that can be directly attributable to stratification/ancestry. They show that it is present in target data from Europe, other continents, even ancient DNA!
28.03.2025 21:53 — 👍 15 🔁 3 💬 3 📌 0
I wrote about how population stratification in genetic analyses led to a decade of false findings and almost certainly continues to bias emerging results. But we are starting to have statistical tools to sniff it out. A 🧵:
28.03.2025 21:53 — 👍 282 🔁 121 💬 8 📌 11
elifesciences.org/articles/48376
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

I find it hard to think of any application where it does not matter. It puts in further question the generalizability of the to samples/patients with different characteristics from the GWAS sample, since axes of confounding may differ.
20.02.2025 13:37 — 👍 1 🔁 0 💬 1 📌 0
BREAKING
The Trump administration is maintaining a funding freeze at the National Institutes of Health (NIH) in defiance of two federal court orders.
The ongoing freeze was confirmed by an NIH official and internal correspondence reviewed by Popular Information.

"One thing is certain: The changes we make ourselves will be healthier than the ones our adversaries demand."
New work for @undark.org:
undark.org/2025/02/06/o...
Think of a polygenic score you care about. Are direct genetic effects driving variation among people in this predictor? Or perhaps other, confounding factors? We at the @arbelharpak.bsky.social & @docedge.bsky.social Labs developed a method to tackle this question. [1/n]
04.02.2025 18:04 — 👍 76 🔁 38 💬 2 📌 1
“The distribution of highly deleterious variants across human ancestry groups”. Preprint with Anastasia Stolyarova and @gcbias.bsky.social: www.biorxiv.org/content/10.1...
02.02.2025 19:00 — 👍 131 🔁 70 💬 1 📌 1Undergrads: apply to NYU's Summer Undergraduate Research Program in Biology!
10 weeks of research in a structured setting, featuring terrific mentors, exciting biology, housing in New York, and a stipend.
as.nyu.edu/biology/outr...
If you’ve been directly impacted by a pause or cancellation of an NIH study session, we @forbes.com would like to talk to you
22.01.2025 22:58 — 👍 2774 🔁 1616 💬 75 📌 35
I am excited to share the first first-author paper of my PhD describing work with Changde Cheng, Mark Kirkpatrick and @arbelharpak.bsky.social has been published at AJHG! We ask if sex-differential gene expression drives sex-differential selection in humans. (1/14)
www.cell.com/ajhg/fulltex...
My lab (aprilweilab.github.io) continues to develop GRG and ARG related methods & more. We are looking for a postdoc to join us.
05.12.2024 17:09 — 👍 4 🔁 5 💬 1 📌 0
A screenshot of a paper on bioarxiv illustrating the lack of blue sky share button!
Would you like to see @biorxivpreprint.bsky.social add a share to blue sky button?! I know I would! Share this post to let @richardsever.bsky.social @erictopol.bsky.social and others at bioarxiv know!
02.12.2024 12:10 — 👍 385 🔁 197 💬 9 📌 6That's simply because there's no one like you. Grateful to have you back!
23.11.2024 04:49 — 👍 3 🔁 0 💬 0 📌 0Three open-rank positions at UT Austin:
Cluster in Advances in Artificial Intelligence and Data Science for Environmental Systems
Apply here: apply.interfolio.com/158908
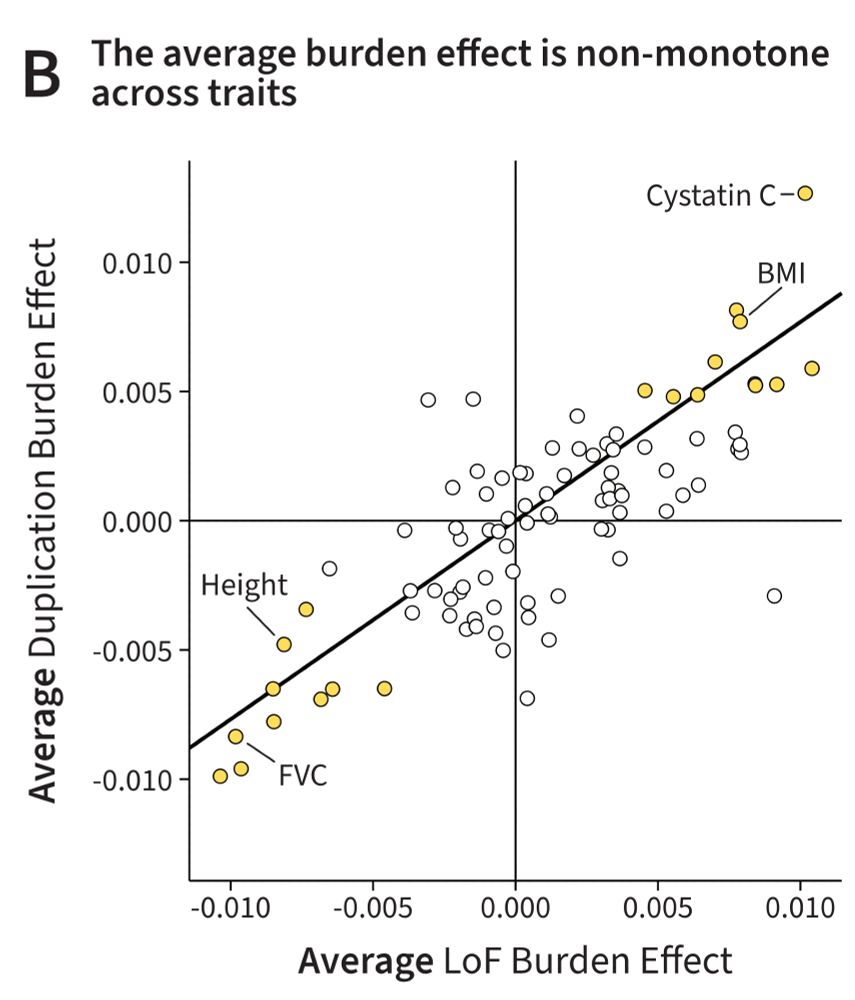
For many traits there is a correlation between the number of duplications or loss-of-function (LoF) mutations someone carries, and their phenotype. Curiously, for most traits, these effects are aligned in the SAME direction. Why?
12.11.2024 06:10 — 👍 46 🔁 25 💬 6 📌 1Thread of some really great works out this week in the world of countering naive genetic thinking (e.g. hereditarianism, genetic determinism, scientific racism)
07.11.2023 21:42 — 👍 40 🔁 26 💬 1 📌 0