
STATGEN 2026 will be in Atlanta @emoryrollins.bsky.social - see more info here: statgen26.emory.edu
05.12.2025 17:01 — 👍 1 🔁 0 💬 0 📌 0@joshweinstock.bsky.social
Assistant Professor in the Department of Human Genetics at Emory University. Statistical genetics and genomics + genetic epidemiology of somatic mosaicism. weinstocklab.org

STATGEN 2026 will be in Atlanta @emoryrollins.bsky.social - see more info here: statgen26.emory.edu
05.12.2025 17:01 — 👍 1 🔁 0 💬 0 📌 0
How do GWAS and rare variant burden tests rank gene signals?
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
Shout to extraordinary collaborators and co-authors on this, Karen Conneely, Cameron Russell, Mitchell Machiela, Marios Arvanitis, Janghee Woo.
For more info, I'm presenting this work on Thursday at #ASHG at 1:45pm, Room 210C

To make results easier to parse, we also release a summary statistics "portal" to view our results, including lots of QC metrics: somatic.emory.edu
We also release lots of code for this, which was all done on the DNA Nexus RAP in a cost effective way.

The IGH and IGL mutations in particular had really large effect sizes, so we then did a GWAS of a combined IGH + IGL phenotype, which resulted in a single hit: GRAMD1B. Remarkably, this is well characterized risk locus for CLL, suggesting that we are converging on CLL relevant biology.
14.10.2025 21:27 — 👍 2 🔁 0 💬 1 📌 0
We then quantified the variance explained on a liability scale of CH to 30 common aging-related diseases. CH explains far more 'liability' scale variance for hematologic malignancies than other classes (as one would expect), though chronic kidney disease appears high here as well.
14.10.2025 21:27 — 👍 0 🔁 0 💬 1 📌 0
New hits are generally indeed "weaker" than most classic CH drivers, suggesting that these are just underneath the "tip of the iceberg". We also replicate the telomere attrition mechanism reported here www.nature.com/articles/s41..., finding that the phenomenon is broader than previously observed
14.10.2025 21:27 — 👍 0 🔁 0 💬 1 📌 0

The strongest hits are indeed strongly enriched for canonical CH genes, but we also find non-coding mutations at FGF1, UGT2B7, DGKB, the TERT promoter, chr17 centromere, and immunoglobulin loci:
14.10.2025 21:27 — 👍 1 🔁 0 💬 1 📌 0
What happens if you search the genomes of ~490K adults for variants that associate with age at blood draw? We actually did this crazy idea, using the UKB WGS (from peripheral blood).
Turns out this 'discovers' classic clonal hematopoiesis drivers and more 🧵👇
www.medrxiv.org/content/10.1...
#ASHG

Exciting updates!!
(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com
Excited for a major milestone in our efforts to map enhancers and interpret variants in the human genome:
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/

I wrote about gene-gene interactions (epistasis) and the implications for heritability, trait definitions, natural selection, and therapeutic interventions. Biology is clearly full of causal interactions, so why don't we see them in the data? A 🧵:
27.08.2025 20:40 — 👍 144 🔁 47 💬 1 📌 6You can find PRSFNN code here: github.com/weinstockj/PRS . It takes in GWAS summary statistics + LD reference panel + annotations, and we compute the posterior using variational inference to make it fast.
A pleasure to build this with @aprilkim.bsky.social and @alexisbattle.bsky.social
We observed similar non-linear effects with AlphaMissense predictions, where low impact coding variants were prioritized, but highly pathogenic variants were not prioritized (presumably because these are so rare in real GWAS).
22.07.2025 21:25 — 👍 2 🔁 0 💬 1 📌 0More compellingly, it also learns non-linear effects wrt to chromatin accessibility - variants in cCREs present in 10-50 cell types were more highly prioritized than variants in cCRE that are present in numerous (> 50) cell types, suggesting a preference for cell/tissue specificity.
22.07.2025 21:25 — 👍 2 🔁 0 💬 1 📌 0In our annotation curation, we included lots of scATAC, cCREs from ENCODE, conservation from Zoonomia, pathogenicity from AlphaMissense, among others. Generally - PRSFNN "learns" that low-frequency SNPs in accessible chromatin are likely to have larger effect sizes (maybe not that surprising).
22.07.2025 21:25 — 👍 2 🔁 0 💬 1 📌 0We connected the SNP annotations to the parameters of the prior distribution on the weights in a novel way with a neural network, so we're calling it Polygenic Risk Scores with Functional Neural Network (PRSFNN). We were excited to see that PRSFNN does well in benchmarks (at least in our hands).
22.07.2025 21:25 — 👍 1 🔁 0 💬 1 📌 0
Really excited to share our new PRS method, developed with @aprilkim.bsky.social and @alexisbattle.bsky.social ! Our approach is to use a lot of recently developed functional annotations to better estimate the weights of the SNPs.
www.medrxiv.org/content/10.1...
Congratulations!
05.06.2025 03:03 — 👍 2 🔁 0 💬 0 📌 0Happy to share our work characterizing functional rare SVs in rare diseases with long-read genome sequencing and transcriptomic outlier data: genome.cshlp.org/content/earl...
26.03.2025 14:30 — 👍 10 🔁 7 💬 1 📌 1Sharing some of our lecture slides on statistical genetics! 🧬
Co-taught with Mike Epstein, Dave Cutler, Karen Conneely, Jingjing Yang, Jian Hu.
Mendelian randomization: weinstocklab.org/lecture_slid...
Biobank scale GWAS methods: weinstocklab.org/lecture_slid...
Hope they're helpful!
We have multiple postdoc positions available in my group at NYU. Join us if you're interested in complex trait genetics and biology. More information about the lab on our website: mostafavilab.org
01.06.2024 13:43 — 👍 19 🔁 15 💬 0 📌 0Specificity, length, and luck: How genes are prioritized by rare and common variant association studies https://www.biorxiv.org/content/10.1101/2024.12.12.628073v1
16.12.2024 10:33 — 👍 46 🔁 24 💬 0 📌 1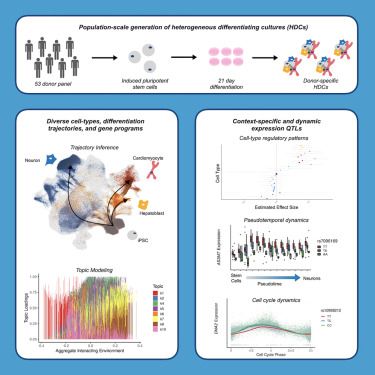
Excited to see our study on genetic regulation in heterogeneous differentiating cultures out in final form!
www.cell.com/cell-genomics/fulltext/S2666-979X(24)00330-6
I'm developing a pipeline to call CHIP mutations in UK Biobank using the DNA Nexus RAP that is fast/cheap/reproducible. Initial results are promising; calls looks reasonable and cost to do this across all of UKB is likely < 500$.
Feel free to DM if of interest.
Great talk from Zeyun Lu on integrating cis-eQTLs with perturb-seq to increase discovery in Mendelian randomization #ASHG2023 !
03.11.2023 15:59 — 👍 14 🔁 2 💬 0 📌 1