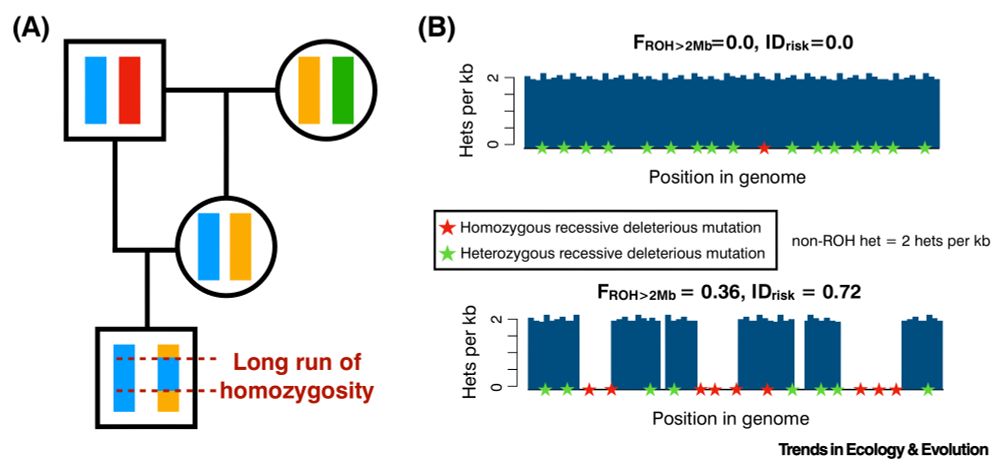
I am thrilled to share this paper outlining some ideas I’ve been thinking about for a little while on a simple but powerful approach for predicting risk of inbreeding depression from long runs of homozygosity and non-ROH heterozygosity. 1/n @klohmueller.bsky.social doi.org/10.1016/j.tr...
04.08.2025 17:10 — 👍 53 🔁 26 💬 1 📌 0
Garud Lab
I am seeking a postdoc for my group at UCLA. We work at the intersection of population genetics x microbiome (garud.eeb.ucla.edu). If interested, please message me!
22.07.2025 17:51 — 👍 86 🔁 100 💬 1 📌 4
Having major FOMO about #SMBE2025 in my hometown… but I’m very proud that my postdoc, Susanna Gutierrez, will be presenting our work on inferring the selection history of adaptive introgression candidates in Peruvians at poster S12-P19. Please visit her during poster session 3 if you're interested!
21.07.2025 06:13 — 👍 19 🔁 4 💬 0 📌 0
Home - ProbGen 2026
Your Site Description
The preliminary program of ProbGen 2026, which will be held at UC Berkeley, is now up: probgen2026.github.io
Sharing on behalf of Rasmus Nielsen who is not on this site. For more see his thread on that other site: x.com/ras_nielsen/...
05.06.2025 20:29 — 👍 20 🔁 13 💬 0 📌 0
Amazing!! Congratulations!!!
03.06.2025 15:43 — 👍 2 🔁 0 💬 0 📌 0
Thank you!
08.05.2025 19:26 — 👍 2 🔁 0 💬 0 📌 0
Thanks Bernard!!
08.05.2025 16:41 — 👍 1 🔁 0 💬 0 📌 0
n/n This was my last postdoc project that finally got completed with tremendous help from my postdoc mentors Kirk and Sriram, two UMich biostats MS students (Jiongxuan Yang and Lingxuan Zhu), and many colleagues and friends in MI and CA (esp. Jazlyn Mooney). Can't thank them all enough!
08.05.2025 15:08 — 👍 1 🔁 0 💬 1 📌 0
6/n Overall, we show that recessive deleterious mutations not only exist on the human genome, but also are not uniformly distributed and are mostly found in regions regulating metabolism and immune functions (our detected regions include the HLA cluster)
08.05.2025 15:06 — 👍 1 🔁 0 💬 1 📌 0
5/n We further investigated the biology of such regions and show they are 1) depleted of haploinsufficient genes, 2) depleted of long ROHs, 3) enriched with UKBB variants with evidence of fitting non-additive model, 4) experienced weaker historical BGS, and 5) more tolerant to LOF mutations
08.05.2025 15:05 — 👍 1 🔁 0 💬 1 📌 0
4/n we applied DominL to 7 non-African pops in 1KG, and show that approx. 3-9% of the human genome is enriched with recessive deleterious mutation! This is important because dominance is very understudied in humans due to many constraints with methodology (eg. can't distinguish h from hs compound)
08.05.2025 15:05 — 👍 1 🔁 0 💬 1 📌 0
3/n our method DominL shows decent power and robustness at finding genomic regions enriched with recessive deleterious muts (or close to fully recessive such as h<0.1; even if the model is trained using two extreme h values) at 1MB resolution, with exceptional accuracy in exon-dense regions
08.05.2025 15:02 — 👍 1 🔁 0 💬 1 📌 0
2/n we developed an ML approach using Neanderthal ancestry as one of the key features and trained 13 ML classifiers with simulations using fully recessive vs fully additive deleterious mutations. After a rigorous performance comparison and feature selection, we picked a winner ML model
08.05.2025 15:00 — 👍 1 🔁 0 💬 1 📌 0
1/n This is a spin-off of some earlier papers showing that recessive deleterious muts can confound adaptive introgression due to heterosis. And we ask a reverse-engineering Q: given the empirical archaic ancestry distribution, can we infer if any genomic region is enriched with recessive mutations?
08.05.2025 14:59 — 👍 1 🔁 0 💬 1 📌 0
Superb!!! Congratulations Fernando!!!!
08.05.2025 00:28 — 👍 1 🔁 0 💬 1 📌 0
Congratulations!! That's a fantastic department and they are lucky to have you!
05.05.2025 18:21 — 👍 1 🔁 0 💬 1 📌 0

Repost w/ correct link!
The Dept of Human Genetics at the Univeristy of Utah is hosting a Rising Stars in Genetics & Genomics postdoc symposium. We are looking to feature postdocs doing cool science! Please self nominate or nominate an excellent postdoc! #ASHGtrainees docs.google.com/forms/d/e/1F...
03.05.2025 04:50 — 👍 23 🔁 26 💬 0 📌 2

Kirk Lohmueller
DominL uses patterns of archaic ancestry to infer degree of dominance. Provides statistical support for the presence of recessive deleterious mutations in the human genome. Up to 15% of the human genome may carry recessive deleterious mutations! 🧬
#HumanEvo25
29.04.2025 15:08 — 👍 11 🔁 6 💬 0 📌 0
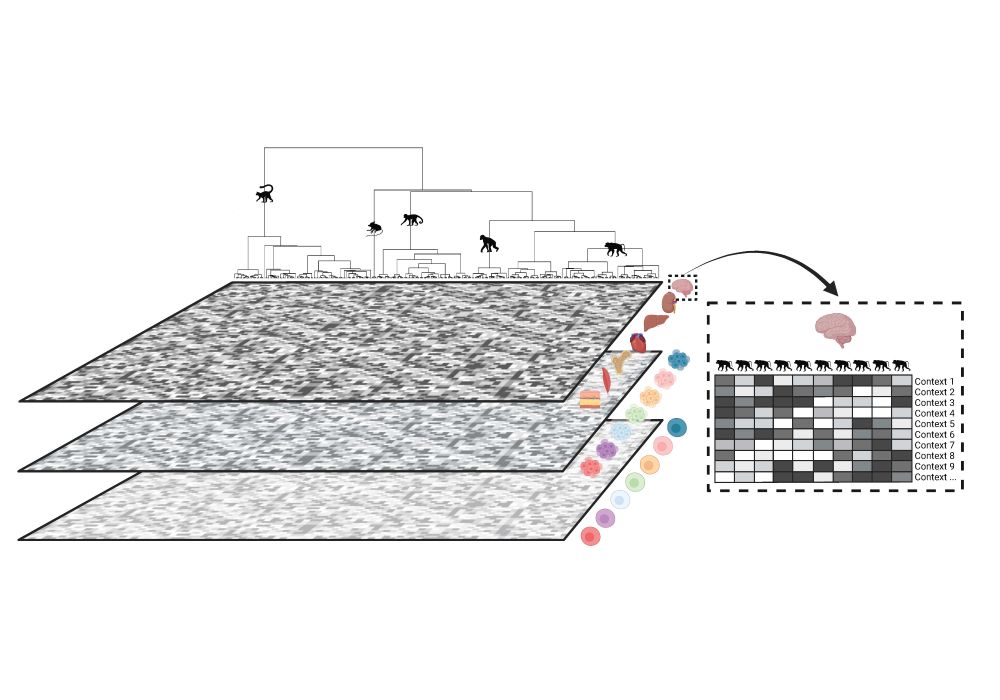
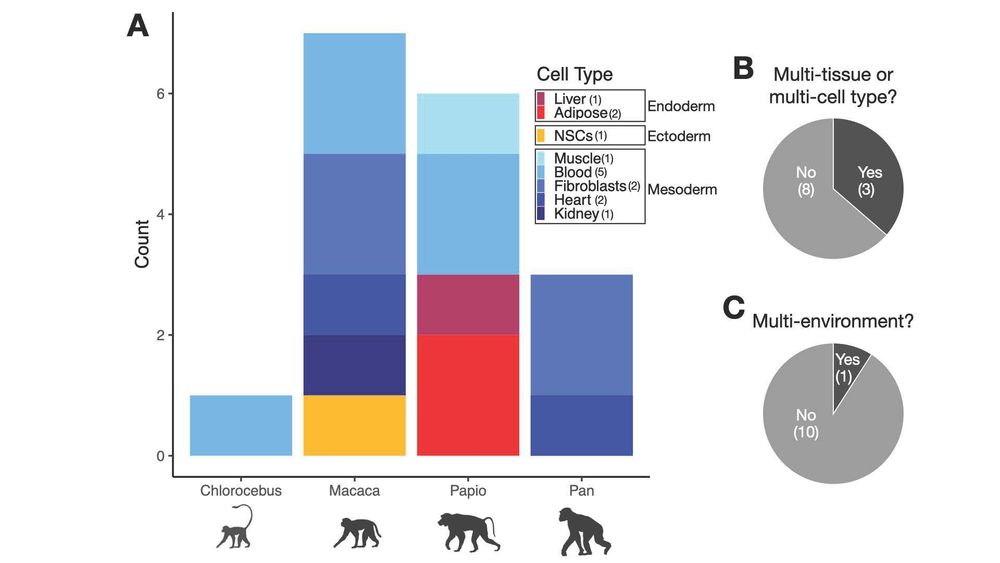
Excited to share: "Addressing missing context in regulatory variation across primate evolution"
arxiv.org/abs/2504.02081
04.04.2025 14:52 — 👍 7 🔁 5 💬 0 📌 0
Congratulations Vince!!
21.02.2025 20:01 — 👍 1 🔁 0 💬 0 📌 0
A Litmus Test for Confounding in Polygenic Scores
Polygenic scores (PGSs) are being rapidly adopted for trait prediction in the clinic and beyond. PGSs are often thought of as capturing the direct genetic effect of one's genotype on their phenotype. ...
Think of a polygenic score you care about. Are direct genetic effects driving variation among people in this predictor? Or perhaps other, confounding factors? We at the @arbelharpak.bsky.social & @docedge.bsky.social Labs developed a method to tackle this question. [1/n]
04.02.2025 18:04 — 👍 76 🔁 38 💬 2 📌 1
Thrilled to have been a part of this study led by @aidaandres.bsky.social that finds evidence of genetic adaptation in chimpanzees to different habitats. Most notably signatures of positive selection in chimpanzees underlie resistance to malaria in humans (GYPA and HBB).
10.01.2025 17:29 — 👍 23 🔁 8 💬 2 📌 0
Population genetics and machine learning
chriscrsmith.github.io
Spartan, Assistant Professor @OhioState. Trying to understand how plants co-evolve with TEs. Author of LTR_retriever, LAI, EDTA, panEDTA, TESS...
Predoctoral researcher at IBE-UPF Barcelona | Biologist & Bioinformatician | Working on human population genetics 🧬
she/her
Prof & Curator @UNM, Museum of Southwestern Biology
NM Game Commissioner
Studies: bird ecology, evolution, genetics, physiology, biogeography, & toxicology
Enjoys: Birding, hunting, fishing, running
Personal account, not representing UNM or State of NM
Postdoc at the University of Zurich | Plant evolutionary genomics 🌱🧬 | Studying supergenes in angiosperms & sex chromosomes in liverworts
Professor passionate about student success & team science #firstgen #Brassica enthusiast walking Dogs of the Plant World
#Agriculture #Food #HigherEd #EduSky #Agsky #SciComm & more
#Soil #Crop #Science ; opinions mine
Instructor at Johns Hopkins University (USA), Associate Researcher at Uppsala University (Sweden), and Editorial Board Member at Communications Biology (UK).
Or just “Li” |
Assist. Prof. @ Plant Bio Michigan State U. |
Also post data visualization |
Lab: https://cxli233.github.io/cxLi_lab/ |
GitHub: https://github.com/cxli233
Assist Prof at UIUC Plant Bio #NewPI. NSF Postdoc UConn EEB. PhD Harvard OEB. Erasmus MEME evobio alumna. She/Her. Lover of ALL plants.
🧪🔬🌱🌾
Development; Evolution; Evo-Devo; Microscopy; Meristems.
Lived in 🇨🇳🇯🇵🇸🇪🇫🇷🇩🇪🇺🇸
www.minyaaa.com
Antropólogo enfocado en evolución humana, genética cuantitativa de fenotipos complejos y el desarrollo de paleogenómica ética y sustentable en México. PhD Candidate en la UMN en el AnthGenLab!!
Assistant Professor @UofUtah Human Genetics | Postdoc @UCSD | PhD @PKU |BS @BNU | Interests: mosaicism in health and development | Lab: https://sites.google.com/view/xiaoxu-yang-lab | Opinions are my own
Moonlighting as a human population geneticists in the Huerta-Sánchez Lab at Brown University, and aspiring speciation geneticists.
PhD candidate in the Edge Lab @USC. Working at the intersection of population genetics and forensic genetics. 🇹🇳
Population and evolutionary geneticist
Institute of Evolutionary Biology (IBE/CSIC-UPF), Spain
Postdoc at Siepel Lab, CSHL
Statistical genetics, phylogenetics, and optimization
Evolutionary biologist, computational biologist, statistician. I like to develop mathematical models of evolutionary process and see how they fit to data. I also like cities where building apartments is legal.
Explorer in Residence, The National Geographic Society https://explorers.nationalgeographic.org/directory/lee-r-berger#c1af6ba3-0859-41e9-a4d6-6b3491785ea8
Professor of Human Evolutionary Biology & Prehistory @CamBioanth @UCamArchaeology - interested in #science, #humanevolution, #prehistory, #palaeontology, #bones #lithics, #fossils, #climatechange, #museums, #collections
Can get distracted by much more 😍