A few places are still available for this training course 🐍🖥️🧬
Don’t miss out, secure your place today and get ready to boost your research! 🚀
🗒️ Registration will be open until Thursday, 28th August 👇
20.08.2025 11:58 — 👍 1 🔁 0 💬 0 📌 0


📣 Big shout-out to Edinburgh Genomics team & Alex Twyford for leading this work on teaching plant genomics! 🌱🧬🖥️🎓
Many thanks to all participants & to ONT for supplying reagents!
#Bioinformatics #Genomics #Training @edgenome.bsky.social @nanoporetech.com
🔗 nph.onlinelibrary.wiley.com/doi/10.1002/...
12.08.2025 15:45 — 👍 1 🔁 1 💬 0 📌 0
📣 Fantastic opportunity to join Joris Veltman’s outstanding group at the Institute of Genetics and Cancer, the University of Edinburgh, advancing research in reproductive genomics and genomic medicine 🚀🖥️🧬
📅 Deadline: 1 Sept
@jorisveltman.bsky.social @uoe-igc.bsky.social @edinburgh-uni.bsky.social
12.08.2025 11:13 — 👍 3 🔁 2 💬 0 📌 0

📢 Ready to level up your coding? Join our next training course:
🐍 Advanced Python for Biologists
📅 1–5 Sept 2025 | 💻 Live online
🔗 Register: genomics.ed.ac.uk/event/advanc...
🧬 A hands-on course for life scientists ready to boost their coding skills in data analysis & tool development.
#Python 🖥️🧬
04.08.2025 17:55 — 👍 6 🔁 1 💬 0 📌 1

Embracing AI with integrity - UK Research Integrity Office
Embracing AI with integrity A practical guide for researchers The UK Research Integrity Office (UKRIO) has launched new guidance to help researchers navigate the research integrity challenges posed by...
New guidance published @ukrio.bsky.social to support responsible AI use in research
This guidance, part of UKRIO’s priority workstream on AI and research integrity, provides a principle-based, researcher-focused resource adaptable across disciplines and technologies.
ukrio.org/ukrio-resour...
27.06.2025 16:25 — 👍 27 🔁 11 💬 0 📌 0

#HiFisequencing with #PacBio is rewriting the rules for dried blood spot analysis.
With Ampli-Fi and SPRQ chemistry, even ultra-low input samples can deliver full, phased genomes that change the game for newborn screening.
Read more: bit.ly/4kbrzm6
27.06.2025 16:27 — 👍 4 🔁 2 💬 0 📌 0

🧬 🖥️ That’s a wrap on our RNAseq Data Analysis course!
🙏 A big thank you to everyone who joined us online last week to explore the world of transcriptomics — from QC and read alignment to gene expression and interpretation.
@edinuni-irr.bsky.social @roslininstitute.bsky.social @uoe-igc.bsky.social
27.06.2025 16:52 — 👍 2 🔁 0 💬 0 📌 1
Single-Cell RNA-seq Data Analysis – Edinburgh Genomics
Join our Single-cell RNA-seq Data Analysis course, running 23–25 June 2025 online!
✅ Process single-cell RNA-seq data
✅ Perform clustering and dimensionality reduction
✅ Identify cell types and gene expression patterns
📅 Book now: genomics.ed.ac.uk/event/single...
11.06.2025 09:13 — 👍 2 🔁 1 💬 0 📌 0

A new compression strategy to reduce the size of nanopore sequencing data. #DataCompression #Nanopore #SequenceData #Bioinformatics #Genomics @nanoporetech.com @genomeresearch.bsky.social 🧬 🖥️
genome.cshlp.org/content/earl...
05.06.2025 07:12 — 👍 6 🔁 3 💬 0 📌 0
Roche has given more details of their SBX NGS technology: it'll be called Axelios, and we know it's aimed at competing against Illumina SBS technology, the technology the San Diego company acquired from Cambridge UK Solexa.
I'll give you a second to catch up with it... Axelios vs Solexa....
01.06.2025 13:43 — 👍 37 🔁 10 💬 2 📌 1
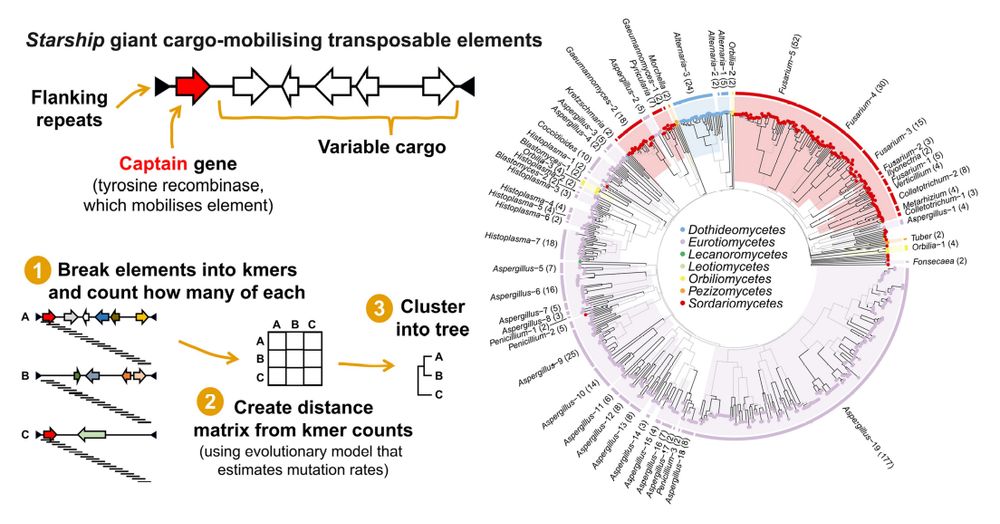
Starship giant transposable elements cluster by host taxonomy using k-mer-based phylogenetics. #TransposableElements #TEs #Kmer #Phylogenetics #Genomics #Bioinformatics #G3 🧬 🖥️
academic.oup.com/g3journal/ad...
02.06.2025 17:06 — 👍 29 🔁 24 💬 0 📌 2

That’s a wrap on our NEB RNA-seq Library Prep Lab Course! 🧪🔬
Big thanks to everyone who took part and made the course a success! 👏
Special thanks to Max Fritsch & Michael Skelly, from New England Biolabs 🦋, for their continued support!
Next course on RNA-seq data 👇
genomics.ed.ac.uk/event/rna-se...
04.06.2025 11:19 — 👍 2 🔁 0 💬 0 📌 1
RNA-seq Data Analysis – Edinburgh Genomics
Want to get to grips with RNA-seq data? Join our online RNA-seq Data Analysis workshop from 16–19 June 2025!
✅ Perform differential expression analysis
✅ Use tools like STAR, DESeq2 & more
🖥️ Hands-on, expert-led, and beginner-friendly.
📅register now: genomics.ed.ac.uk/event/rna-se...
04.06.2025 09:14 — 👍 1 🔁 0 💬 0 📌 0

📣 We still have a couple of spots left in our online training courses next week!
🧬 Linux for Genomics (9–10 June)
👉 genomics.ed.ac.uk/event/introd...
📊 R for Biologists (11–13 June)
👉 genomics.ed.ac.uk/event/r-for-...
Don’t miss out - last places available!
03.06.2025 14:29 — 👍 1 🔁 0 💬 0 📌 0
📣 There are still a few places available on our upcoming #RNASeq library prep Course, running next week, 28–29 May, at Edinburgh Genomics. 🧬🧪
⏳ Don’t miss out – registration closes soon!
👉 genomics.ed.ac.uk/event/rna-li...
#Sequencing #Genomics #RNAseq #LabTraining
21.05.2025 16:29 — 👍 1 🔁 0 💬 0 📌 0

🔬 That’s a wrap on our Metabarcoding & Metagenomics workshop!
4 days of hands-on tutorials, microbiome insights, and plenty of Qiime2, NanoClust, and HUMAnN action 🧫💻
Huge thanks to our amazing participants & instructors!
#bioinformatics #metagenomics #EdGenTraining
12.05.2025 12:17 — 👍 4 🔁 0 💬 2 📌 0

It was great to welcome colleagues from the
Sarawak Infectious Disease Centre this week!
🔬They visited Edinburgh Genomics, with productive discussion around genomics infrastructure, data generation, and long-read sequencing
🌏 Always a pleasure to connect with international teams driving innovation
09.05.2025 13:08 — 👍 2 🔁 2 💬 1 📌 0

Join our 2-day RNA Library Preparation Lab Course at Edinburgh Genomics:
📅 28–29 May 2025
📍 Ashworth Labs, University of Edinburgh
🧪 Use @NEBiolabs NEBNext UltraExpress kits
🧬 Option to bring your own sample!
👉 genomics.ed.ac.uk/event/rna-li...
📸 See our last course in action!
07.05.2025 10:49 — 👍 1 🔁 1 💬 0 📌 2

🚨 Our Intro to #Python course starts next week and there’s still space!
🧬 Perfect for #biologists & #genomic scientists
💻 🖥️Learn to automate tasks, analyse data & build #bioinformatics pipelines
📍 In-person
📅 Plus #R & #Linux courses in June!
🔗 Sign up: genomics.ed.ac.uk/training
06.05.2025 12:23 — 👍 4 🔁 0 💬 0 📌 0

That’s a wrap on our first-ever Full Length 16S rRNA @pacbio.bsky.social Sequencing & Bioinformatics course! 🧪💻
From lab bench to species-level insight, participants processed their own data through QC, DADA2, taxonomic annotation & more, to create figures like this! 🧬👇
23.04.2025 10:12 — 👍 7 🔁 2 💬 0 📌 0

That’s a wrap on last week’s #Snakemake ! Thank you to our participants for their enthusiasm—hope you’re ready to supercharge your workflows! 🐍🖥️🧬
Next week 16S lab and bioinformatics: genomics.ed.ac.uk/event/full-l...
Then long read bioinformatics April 28 - May 1
genomics.ed.ac.uk/event/introd...
03.04.2025 11:28 — 👍 3 🔁 0 💬 0 📌 0
Why does this matter?
Choosing the right extraction method is essential for getting high-quality sequencing data. Our team has tested these approaches to ensure the best possible results!
Have an extraction method you swear by? Let’s discuss!👇#Genomics #DNAExtraction
🧵5/5
28.03.2025 16:47 — 👍 1 🔁 0 💬 0 📌 0
⚠️ CRUCIAL WARNING ⚠️
❌ DO NOT use phenol-chloroform extractions if you're planning to sequence with PacBio - it can interfere with sequencing chemistry and lead to poor results.
@pacbio.bsky.social
🧵4/5
28.03.2025 16:47 — 👍 1 🔁 0 💬 1 📌 0
Long-read sequencing (ONT/PacBio) needs high molecular weight (HMW) DNA for optimal read length.
Our top choices:
🔹 NEB Monarch HMW DNA
🔹 promega Wizard HMW DNA
@nebiolabs.bsky.social @promegacorporation.bsky.social @promegauk.bsky.social
🧵3/5
28.03.2025 16:47 — 👍 2 🔁 0 💬 2 📌 1
Director of UCSF
Center for Advanced Technology. cat.ucsf.edu. Interests in genomics, metagenomics, automation, molecular biology.
Genomics Core Director at Van Andel Institute. VAIGS professor. Runner | Baker | Kid & Pet Wrangler
A tall, sturdy creature fond of industry and DNA sequencing.
By day, I’m unraveling the mysteries of genomes, managing the NGS facility passionately. By night, I’m a multitasking mom cherishing time with my kids. As an activist and union member, I channel my energy into making a difference. Opinions here are my own.
Spatial Tech/CITE-seq/TotalSeq/Slide-seq/Slide-tags Afficionado, space 🚀 enthusiast and nerd for new tech!🐼 Curio Bioscience - Director of Business Development
Find my violin videos here :)
http://www.youtube.com/byeung
Vienna BioCenter Scientific Training Unit, we share opportunities that exist at the Vienna BioCenter (VBC), which include our PhD, Post Doc and Summer School programs. https://training.vbc.ac.at/
Clinician-scientist / precision medicine / cancer immunology / bioinformatics / MD, PhD
@Netherlands Cancer Institute / ESMO precision medicine working group
Trying to have fun discovering stuff to help patients.
Opinions are my own.
🇸🇪 Sweden's national center for molecular biosciences
👩🔬 Infrastructure with unique technologies and expertise
👥 Research community
🥼 Life science 🌱
💻 Often data-driven
https://www.scilifelab.se/
https://www.linkedin.com/company/scilifelab/
Functional Genomics Center Zurich (FGCZ) is a joint platform of the ETH Zürich and the University of Zurich. #genomics #proteomics #metabolomics
Check us out on LinkedIn: https://tinyurl.com/fgczlinkedin
Genomic core director broadly interested in science, art, and kid/pet stories.
🧬 National Genomics Infrastructure hosted by @scilifelab.se
🤖 State-of-the-art technology and technology development
👩🔬 Expertise and support for applying genomics in life sciences
https://ngisweden.scilifelab.se
43 years of academic biotech DFCI/HMS ending with genomics 🧪 🧬
Scientist, community builder, parent. Enabling the future generation.
DNA and CRISPR enthusiast ✂️🧬
ABRF President 25-26
Support your local Shared Research Resource
Director Duke Sequencing & Genomics Tech / Asst Prof MGM / cis-regulatory evolution / DEI / Mama / she | her / daughter of immigrants / wearer of many hats, owner of one head / posts my own
Michele Banks
Artist inspired by science
Friend to squirrels
https://www.etsy.com/shop/artologica/

























