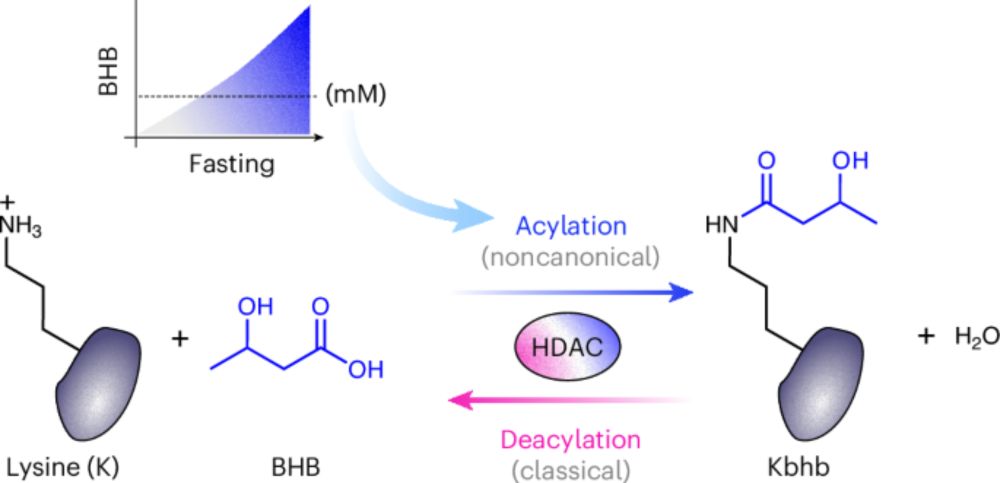
10.02.2026 14:11 — 👍 57 🔁 13 💬 4 📌 1
10.02.2026 14:11 — 👍 57 🔁 13 💬 4 📌 1
QB3 MacroLab – QB3 Berkeley
When I was in @hitenmadhani.bsky.social lab, we used a blend of lab-purified taq and Pfu to do colony PCRs on Cryotococcus that was robust. You can purchase Phusiom from the UCB MacroLab qb3.berkeley.edu/facility/qb3...
28.10.2025 20:06 — 👍 1 🔁 0 💬 1 📌 0


Awesome turnout @standupforscience.bsky.social SF today! The Oregon Trail inspired sign was one of my favorite!
08.03.2025 00:32 — 👍 10 🔁 2 💬 2 📌 0
Yup, but any of the current single cell antibody panels should work.
28.02.2025 14:44 — 👍 0 🔁 0 💬 0 📌 0
Hi John, we do think it's possible to capture protein information using tagged antibodies that have polyA barcodes or if using direct capture ADTs, spike in a splint oligo.
28.02.2025 05:06 — 👍 0 🔁 0 💬 1 📌 0

Welcome to the Bluesky account for Stand Up for Science 2025!
Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence
12.02.2025 17:04 — 👍 11504 🔁 5438 💬 291 📌 670
Just look at these paragraphs from the AP story.
Hegseth called Brown unqualified solely because he's Black. Then they fired him... and replaced him with a white guy so indisputably unqualified that he requires a Presidential waiver.
This is what "merit" means to them.
apnews.com/article/trum...
22.02.2025 01:53 — 👍 15878 🔁 5925 💬 463 📌 319
Can I give some unsolicited advice? When communicating on this catastrophe, don’t lead with “I know the NIH is inefficient and needs serious reform, but…”. You are undercutting the entire message and buying into the frame of the arsonists. This is not a scientist audience. No “to be fair” points
08.02.2025 05:31 — 👍 76 🔁 11 💬 4 📌 0
I’m reaching out to my vendors to push them to use their lobbying infrastructure to point out the devastating effects the NIH change will have on the USA’s R&D engine.
08.02.2025 04:50 — 👍 16 🔁 4 💬 0 📌 0
all y'all academics who have been naively convinced IDC only goes to useless sub deans are about to find out. they are going to claw the real costs of research out of your unchanged direct costs.
07.02.2025 23:47 — 👍 88 🔁 23 💬 9 📌 4
Indirect rates are not determined by NIH. Instead, they are the product of elaborate negotiations between each institution and the “cognizant federal agency” according to an Office of Management and Budget document, Circular A-21.
1/n
08.02.2025 00:51 — 👍 455 🔁 122 💬 6 📌 14
Congratulations Chris!
05.02.2025 01:32 — 👍 1 🔁 0 💬 0 📌 0
No worries Andrew. Thank you for the responses, this info is helpful. Can’t wait to try DV on our future system!
01.02.2025 04:58 — 👍 2 🔁 0 💬 0 📌 0
Hi Andrew, we are spec’ing out a server. It seems DV utilizes extensions on Intel CPUs, so we’ll go with them. Any thoughts on the L4 vs L40S. Is there a possibility DV will require >16GB GPU memory in the near future?
Thanks!
25.01.2025 22:10 — 👍 0 🔁 0 💬 1 📌 0
We only upgraded one of our systems as a “just in case”. Our update required an engineer visit so that might also play a role?
23.12.2024 22:48 — 👍 1 🔁 0 💬 1 📌 0
We are currently offering 10X GEX lane queues (28x10x10x90). If there is enough interest for a PE50/10X ATAC (51x12x24x51), we will start that as well.
23.12.2024 03:45 — 👍 2 🔁 0 💬 0 📌 0
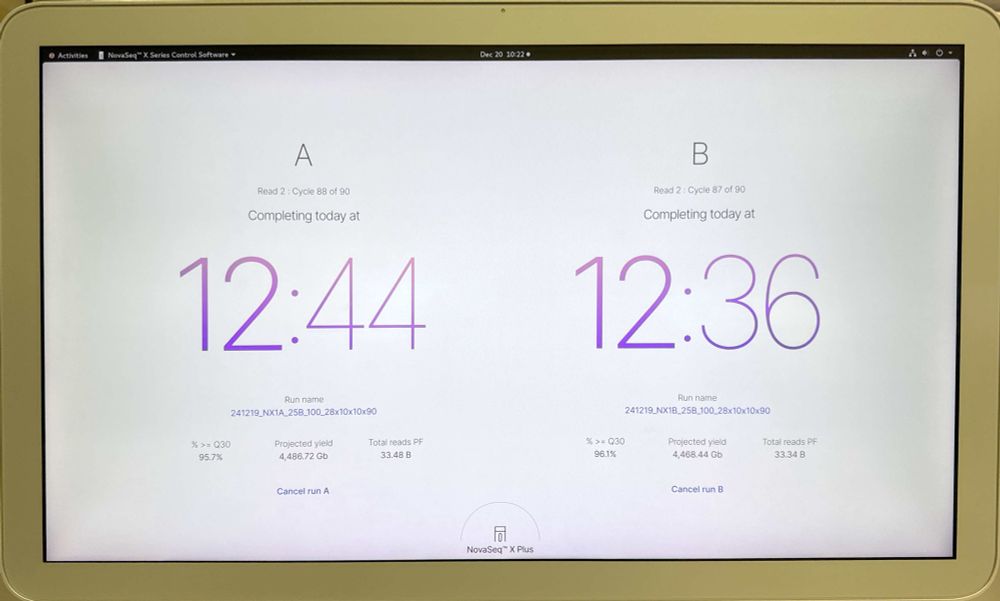
We just upgraded one of our Illumina NovaSeqX’s to enable the new 25B 100 and 200 cycle kits. New software is supposed to slightly raise PF% and we are seeing that. 10X GEX libraries previously generated 32B reads per flow cell. We seeing over 33B, almost 67B reads in 24 hours for a dual FC run!
23.12.2024 03:44 — 👍 9 🔁 2 💬 2 📌 0
Some preps recommend different cycles (10X SC and for Visium a qPCR titration). With clinical metagenomic samples, we are not comparing across samples so no worries there.
We’ll look at for bulk-RNA-seq experiments but even with constant cycles, the n6 will give you quant and pooling info.
18.12.2024 16:42 — 👍 2 🔁 0 💬 0 📌 0
UCSF users, we will have a training to be scheduled the second week of January. Details will go out on our listserve and posted on our website.
18.12.2024 05:22 — 👍 2 🔁 0 💬 1 📌 0
The iconPCR solves these issues. Samples will be within 2-fold concentration of each other and the software uses the fluorescence level to calculate relative concentrations and pooling volumes to further normalize pools. You get a pool that’s ready to go on your production scale sequencing runs.
18.12.2024 05:22 — 👍 2 🔁 0 💬 1 📌 0
This works fairly well but adds time and cost and some samples are amplified too little, and others too much, resulting in adapter dimer carryover in bubble products.
18.12.2024 05:22 — 👍 2 🔁 0 💬 1 📌 0
We see many applications but our first one will be metagenomics from clinical samples where inputs vary widely. To Amie this high throughput, we have been pooling and equal volume of many samples after PCR, doing a cleanup on the pool, running a MiSeq nano, and repooling based on read distribution.
18.12.2024 05:22 — 👍 1 🔁 0 💬 1 📌 0
The system will place each sample at a low temp hold once they have reached a user-defined set point. If anyone used the old Kapa Real Time NGS amp kit, this allows you to do this automatically with your choice of PCR mix. Just add a little bit of SYBR Green.
18.12.2024 05:22 — 👍 1 🔁 0 💬 1 📌 0
Introducing iconPCR
Discover simplicity and innovation combined in our groundbreaking icon PCRTM technology. Stay tuned as we usher in a next paradigm in genomics solutions.
We just got our n6 Tec iconPCR installed. The system monitors NGS library amp with real time functions but each of the 96 wells is independently temperature-controlled and monitored. This is super helpful when you have varying inputs. www.n6tec.com
18.12.2024 05:22 — 👍 15 🔁 2 💬 5 📌 1
The new MiSeq i100 should be good to check pooling for the NovaSeqX. Catherine's group is seeing good consistency between the two platforms, but from first glance, it does appear the MiSeq i100 is more consistent with the older iSeq (blue and orange lines).
16.12.2024 19:17 — 👍 4 🔁 1 💬 0 📌 0
Hi Andrew, any benchmarking on different GPUs? Particularly interested comparing the L40S and L4. Thanks!
06.12.2024 00:17 — 👍 0 🔁 0 💬 1 📌 0
150pM by our custom ddPCR assay we’ve been using for the past decade. Taking loading concentration with a grain of salt - as you know, different methods and users can result in different quants, especially our assay which uses a 10^7 dilution.
03.12.2024 22:52 — 👍 3 🔁 0 💬 0 📌 0
Music, walking, ventilation.
Research fellow
Chair of the CIBSE natural ventilation group
We are n6 and we bring you the iconPCR–the first and only thermocycler that intelligently adjusts amplification in real time—so you get normalized, sequencing-ready libraries in one automated step.
https://www.n6tec.com/
Associate Professor
DFCI & HMS
Research Engineer in the Technology Innovation Lab @ NYGC. I like to repurpose sequencers for spatial omics applications.
Husband, Father, Grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science, Stand Up for Science advisor, Shenanigator, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Dad, husband, President, citizen. barackobama.com
assistant professor @mskcancercenter.bsky.social
clareaulab.com
A world leader in health care, we're proud to rank among the nation's top hospitals. Follow our children's hospitals @ucsfchildrens
Product manager @PacBio / RNA / bioinformatics / aerialist & pole dancer / opinions are my own and do not reflect views of my employer
New York Times bestselling author. Former All Things Considered, NPR, LA Times, NME, McSweeney’s, Stanford University. Singer in this one band.
Pediatric neuro-oncologist, Asst Prof U-Michigan | Cancer researcher | RNA & ribosome enthusiast | Broad Institute, DFCI alum | book & music & tea lover | Dad | Views are mine (he/him). https://prensnerlab.org
Original and probably disagreeable perspectives on genomics. Not sure when I'm joking and when I'm being serious. Opinions my own, not the herd’s.
Leading developer of CUT&RUN and CUT&Tag chromatin mapping assays and other tools for chromatin research. Follow us for the latest news in epigenetics.
physician-scientist Assistant Professor @ucsf. #newPI stemcellbio+RNA/omics scientist | liver pathologist | founder @MDPhDEquity.bsky.social | BWF CAMS | ASCI YPSA | alum @ucsf @DukeMSTP @Stanford | http://csangokoya-mdphd.com | sangokoyalab[dot]ucsf.edu
Chemical biologist. Currently working on spatial omics for 10x Genomics.
Our laboratory seeks to understand how chromosome structure relates to genome functions
Professor and Chair @UCSF/@UCSF_BTS - dynamic structural biology and open science - (he/him) - fraserlab.com
Computational biologist & blogger
 10.02.2026 14:11 — 👍 57 🔁 13 💬 4 📌 1
10.02.2026 14:11 — 👍 57 🔁 13 💬 4 📌 1