Congrats Grace, Max, and team! We in the Collins lab are looking forward to diving into the paper!
03.07.2025 04:54 — 👍 0 🔁 0 💬 0 📌 0
Vertebrate retrotransposons are the future of gene therapy. But how do they insert their genes? 🔥🔥
Thrilled to share our new work now published with Kathy Collins, @nogaleslab.bsky.social @berkeleymcb.bsky.social where we investigate this with #cryoEM & biochemistry in 🧪 and cells! #RNAsky #TEsky
23.06.2025 16:19 — 👍 68 🔁 21 💬 2 📌 2
Structures of vertebrate R2 retrotransposon complexes during target-primed reverse transcription and after second-strand nicking pubmed.ncbi.nlm.nih.gov/40540573/ #cryoEM
21.06.2025 15:48 — 👍 4 🔁 4 💬 0 📌 1
An NIH staffer reacts to today's ruling:
"I'm looking forward to the day that we are so slammed with work trying to reinstate everything that we had to terminate illegally — I'll work 24/7 to make that happen if I can."
16.06.2025 22:24 — 👍 1117 🔁 297 💬 9 📌 15
A big chunk of my PhD work with the Collins lab is now on bioRxiv!
14.05.2025 03:43 — 👍 6 🔁 3 💬 0 📌 0

Stanford.Berkeley.UCSF Next Generation Faculty Symposium
Are you a postdoc/grad student preparing to launch a faculty search? Do you have a track record of excellence in research, leadership, mentorship & community engagement? Apply to the 2025 Next Generation Faculty Symposium: www.berkeleystanfordnextgensymposium.com! Pls repost! (1/3)
12.05.2025 18:06 — 👍 13 🔁 19 💬 1 📌 0
Lineage-Specific Evolution, Structural Diversity, and Activity of R2 Retrotransposons in Animals https://www.biorxiv.org/content/10.1101/2025.05.05.652312v1
10.05.2025 01:31 — 👍 8 🔁 4 💬 0 📌 2

Kathleen Collins elected to the National Academy of Sciences
Congrats to MCB's Kathy Collins on her election to the National Academy of Sciences! 👏🎉https://www.nasonline.org/news/2025-nas-election/
29.04.2025 21:34 — 👍 8 🔁 3 💬 0 📌 0

Deep sequencing of yeast and mouse tRNAs and tRNA fragments using OTTR
It is finally (FINALLY) out! We characterized Kathy Collin's super cool OTTR protocol for tRNA and tRF cloning, showing that it is comparable (mim-tRNA-seq, maybe nano-tRNA -seq and LIDAR) or superior (everything else) to the best protocols in the literature.
elifesciences.org/articles/77616
28.04.2025 00:33 — 👍 19 🔁 8 💬 0 📌 1
This is so cool! Congrats Emily!!
25.04.2025 02:46 — 👍 1 🔁 0 💬 1 📌 0

Jennifer Doudna speaking at Stand Up for Science in Berkeley

crowd at Stand Up for Science in Berkeley

crowd at Stand Up for Science in Berkeley

crowd at Stand Up for Science in Berkeley
Thanks to out @ucberkeleyofficial.bsky.social community for joining us at Stand Up For Science! @jenniferdoudna.bsky.social spoke about how NIH funding supported her PhD & NSF funding supported her development of #CRISPR genome editing. Federal funding is needed for life-saving science!
08.03.2025 00:03 — 👍 91 🔁 28 💬 0 📌 2
What's better than 1 deep mutational scanning (DMS) library? 2! In a new pre-print @brittneywthornton.bsky.social and @rfw.bsky.social et al. map the mutational landscape of ISDra2 TnpB protein and reRNA and leverage these datasets to engineer highly active variants (1/9)
26.02.2025 18:43 — 👍 15 🔁 7 💬 1 📌 1
🚀 Meet Eliel Akinbami, a Stanford BioE PhD student in @pollyfordyce.bsky.social Lab! Their research has led to HT-MEK (High-Throughput Microfluidic Enzyme Kinetics), a groundbreaking technique that condenses years of enzyme research work into just weeks. 🔬 @stanford-chemh.bsky.social #stanfordbioe
04.02.2025 18:34 — 👍 9 🔁 3 💬 0 📌 1
I was stunned by this work from Kathleen Collins' lab (Berkeley) when I heard her present this at a FASEB meeting!
What a crazy innovative idea: Adapting the R2 retrotransposon to efficiently insert any custom transgene directly into RIBOSOMAL DNA ARRAYS! 🤯🤯🤯
The applications are endless....
05.02.2025 22:55 — 👍 18 🔁 5 💬 0 📌 0
My favorite!!
28.12.2024 02:19 — 👍 1 🔁 0 💬 0 📌 0

Changing fitness effects of mutations through long-term bacterial evolution
Predictable and parallel changes occur in the fitness effects of mutations in Escherichia coli over 50,000 generations.
A new LTEE paper is out. They don't use the term "adjacent possible," but the idea is there.
As populations accumulated mutations, "some evolutionary paths that were inaccessible to the ancestor became accessible to the evolving populations, while others were closed off."
🦋🦫🌱🐋🧪 #philsci #evobio
01.02.2024 23:18 — 👍 17 🔁 8 💬 2 📌 0

A big step forward in the science of aging: organ-specific tracking via plasma proteins
www.nature.com/articles/s41...
20% of people assessed had accelerated aging in 1 organ, 1.7% multi-organ
06.12.2023 20:50 — 👍 41 🔁 16 💬 0 📌 1

Codon optimality modulates protein output by tuning translation initiation
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
Spurred on by ribosome profiling and other cool methods, translation elongation has grown into a whole field with new surprises all the time (and new relevance for vaccine mRNAs!). Here’s our latest on why synonymous codons aren’t all the same. (Thread) www.biorxiv.org/content/10.1...
29.11.2023 07:07 — 👍 43 🔁 13 💬 1 📌 1
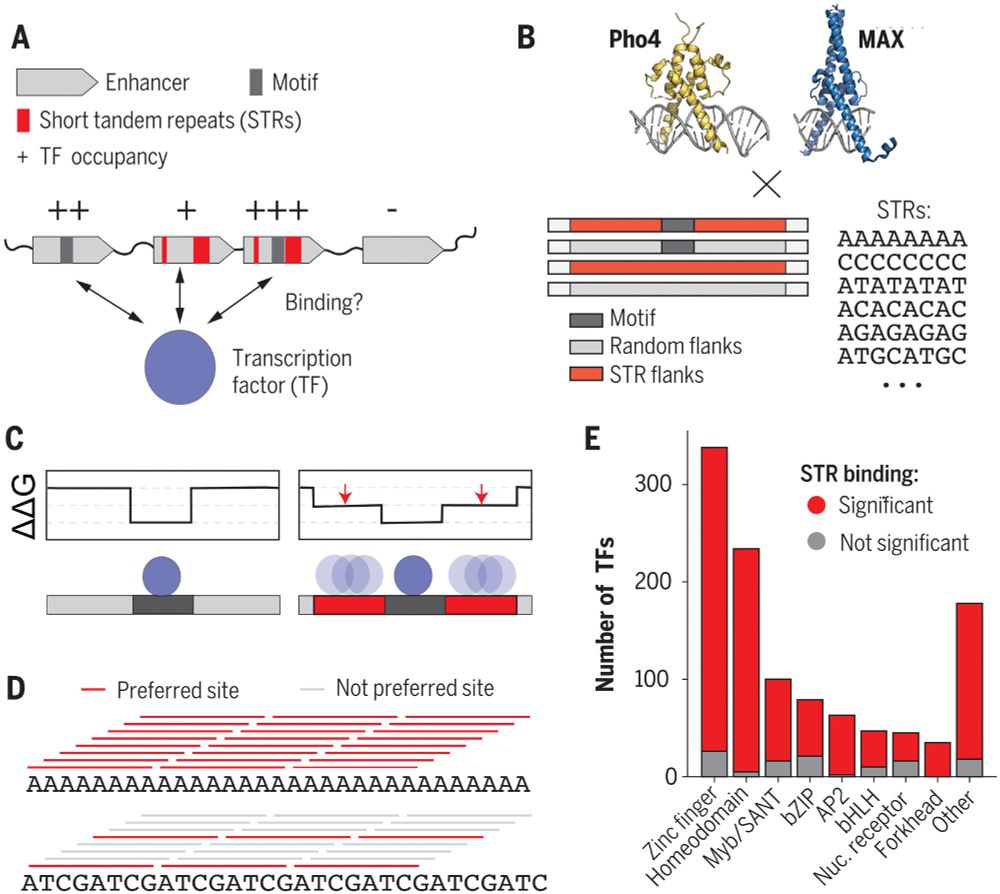
Short tandem repeats (STRs) are common in eukaryotic genomes. Horton et al. in Science show transcription factors (TFs) directly bind STRs by a additive model - low-affinity bindings amount to large effects suggesting STRs tune TF binding and regulate gene expression bit.ly/add1250 bit.ly/adk2055
03.10.2023 15:18 — 👍 0 🔁 1 💬 0 📌 0

I see this all the time - you don't need to wait for a journal to ask you to review - JUST REVIEW THE PREPRINT and post your review!
We do it all the time: fraserlab.com/reviews
The majority of the time, a journal asks us to include the review in their process AFTER we post!
04.10.2023 15:22 — 👍 13 🔁 7 💬 0 📌 0
And biggest thanks of all to Polly @pollyfordyce.bsky.social who saw the promise of this project from the beginning and believed in me every step of the way! I learned so much from you! (11/11)
21.09.2023 20:03 — 👍 1 🔁 0 💬 0 📌 0
This project would not have been possible without the awesome interdisciplinary team! Amr Alexandari, Michael Hayes, Emil Marklund, Julia Schaepe, Arjun Aditham, Peter Suzuki, Nilay Shah, Avanti Shrikumar, Ariel Afek, Raluca Gordan, Will Greenleaf, Julia Zeitlinger, and @anshulkundaje.bsky.social
21.09.2023 20:02 — 👍 2 🔁 0 💬 1 📌 0

STRs are a case study in the importance of sequence context and low-affinity binding! We hope our work inspires others to think beyond TF motifs! (9/11)
21.09.2023 20:01 — 👍 0 🔁 0 💬 1 📌 0

By analyzing published protein-binding microarray data, we predict that 90% of eukaryotic TFs bind at least one type of STR above background, suggesting that STRs (which are enriched in enhancers!) function as regulatory sequences. (8/11)
21.09.2023 20:00 — 👍 1 🔁 0 💬 1 📌 0

Kinetic measurements and modeling work from Emil Marklund and Julia Schaepe show that STRs primarily alter apparent association rates (k_on), perhaps affecting TF target search. (7/11)
21.09.2023 20:00 — 👍 0 🔁 0 💬 1 📌 0

De novo distillation of thermodynamic affinity from deep learning regulatory sequence models of in v...
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
Teaming up with Amr Alexandari in @anshulkundaje.bsky.social's lab, we trained neural net models on ChIP-seq data to show that the same repeats altering in vitro binding alter TF occupancy in cells.
Check out www.biorxiv.org/content/10.1... for more! (6/11)
21.09.2023 19:59 — 👍 7 🔁 2 💬 1 📌 0

What part of the TF is responsible for recognizing STRs? We made truncation mutants and single-residue substitutions of TFs and show that the DNA-binding domain directly recognizes STRs. (5/11)
21.09.2023 19:57 — 👍 1 🔁 0 💬 1 📌 0

Using partition function models from statistical mechanics, we show that STRs are low-affinity sites that maximize the number of binding microstates.
And we can apply these models to vastly improve binding predictions for any arbitrary TF and DNA sequence! (4/11)
21.09.2023 19:57 — 👍 0 🔁 0 💬 1 📌 0

Using high-throughput microfluidic binding assays, we showed that STRs can alter transcription factor binding affinities to a surprisingly large degree, up to 70-fold!
And here's the weird part: the STRs don't need to resemble high-affinity motifs to affect binding. (3/11)
21.09.2023 19:56 — 👍 2 🔁 0 💬 1 📌 0
Postdoc at Stockholm University. TEs, epigenetics, bioinformatics, small RNA and Drosophila 🪰!
artemilin.dev
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Scientist at IMP in Vienna. Excited about gene expression regulation and its encoding in our genomes - enhancers, transcription factors, co-factors, silencers, AI.
Graduate Student @ Princeton Chemistry
M. Chang Lab
NSF Graduate Research Fellow
Stanford '20
genes, circuits, and behavioral evolution
Assistant Professor of Neural Science & Biology at NYU
he/him
I’m a computational biologist and biochemist working on synthetic tumor genome generation 💻🧬 Postdoctoral researcher at University of Toronto (UofT) and Ontario Institute for Cancer Research (OICR)
Postdoctoral Researcher at UPenn in the lab of Michael Lampson. Constantly amazed by evolutionary mechanisms in all aspects of biology. Chromosome segregation aficionado.
Professor of Creative Pedagogies | Poet | Game Designer | Slow AI
#SciComm #HigherEd #Poetry #GenAI
https://theslowai.substack.com/
https://linktr.ee/sam.illingworth
Virus spread and evolution + connection to behavior.
@Scripps Research, CA.
Philosophy of Science - University of Valencia
http://saulpe4.blogs.uv.es/
Whitehead Institute and Department of Biology, MIT. Lover of cell biology and cell division. Aspiring to do good science and do good.
Baylor College of Medicine
History & Philosophy of Science @ Boston University
mohnesorgehps.com
PhD student | Mannervik lab | Stockholm University | developmental epigenetics and chromatin of Drosophila| histone acetylation and methylation
Postdoc in the Zhang lab @ Broad Institute. Formerly in the Elowitz lab @ Caltech.
Née Ke-Huan Chow.
I ❤️ SynBio and cats.
Research scientist at @GoogleDeepMind passionate about AI, genomics and biology.
Genome Scientist. Studies how DNA makes humans, mice, plants, microbes. At Lawrence Berkeley National Lab and Joint Genome Institute. Views are my own.






















