
Such a fantastic resource by @kuhlwilm.bsky.social's lab. We’re using this catalog in half our projects
www.nature.com/articles/s41...
@david-gokhman.bsky.social
Asst Prof @WeizmannScience. Exploring human evolution through the lens of gene regulation🧬 Still rooting for Neanderthals & Denisovans💀❤️ gokhmanlab.com

Such a fantastic resource by @kuhlwilm.bsky.social's lab. We’re using this catalog in half our projects
www.nature.com/articles/s41...
Another truly amazing work by the Capellini lab
28.08.2025 06:41 — 👍 5 🔁 1 💬 0 📌 0It was a true pleasure to work on this with @nadavmishol.bsky.social, @lirancarmel.bsky.social
, Gadi Herzlinger, Uzy Smilansky, and Yoel Rak

Our approach was based on gene regulatory phenotyping, which predicts the direction, rather than magnitude, of phenotypic difference - a much more attainable goal. If you're interested in why and how this approach works, see: www.nature.com/articles/s41...
27.08.2025 13:05 — 👍 3 🔁 0 💬 1 📌 0This approach opens a new way to connect genetics & fossils, helping classify debated human remains and refine our evolutionary tree.
27.08.2025 13:05 — 👍 1 🔁 0 💬 1 📌 0
Credit: Maayan Harel
Denisovans or their close relatives identified in the fossil record using a gene regulatory phenotyping method. Now out:
www.pnas.org/doi/10.1073/...
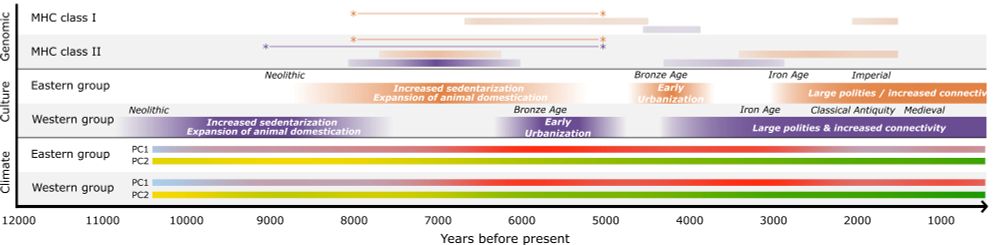
🧵 Excited to share a new preprint from our lab looking into signals of disease-burden in aDNA (doi.org/10.1101/2025...)! It has long been speculated that transition to sedentary, agricultural and urbanized lifestyle increases disease burden, but can we see this in aDNA?
01.08.2025 17:55 — 👍 18 🔁 6 💬 2 📌 0Overall, we suggest that substantially more phenotypic information can be inferred from genetic data than previously appreciated. Had a great time working on this with @gilig.bsky.social @shaicarmi.bsky.social and Keith Harris
26.07.2025 17:14 — 👍 3 🔁 0 💬 0 📌 0Importantly, unlike quantitative phenotypic predictions (e.g., based on polygenic scores), we show that our qualitative approach circumvents one of the biggest problems in population genetics – the limited transferability of genotype-phenotype association data across populations
26.07.2025 17:14 — 👍 1 🔁 0 💬 1 📌 0This estimator can be applied to comparisons of individuals from the same family, same population, different populations, or even different species
26.07.2025 17:14 — 👍 0 🔁 0 💬 1 📌 0Our method works even when genotype-to-phenotype information is limited. For example, we show that even in phenotypes where known SNPs explain only 3% of phenotypic variation, you can often predict with >90% accuracy who has the higher phenotypic value
26.07.2025 17:14 — 👍 1 🔁 0 💬 1 📌 0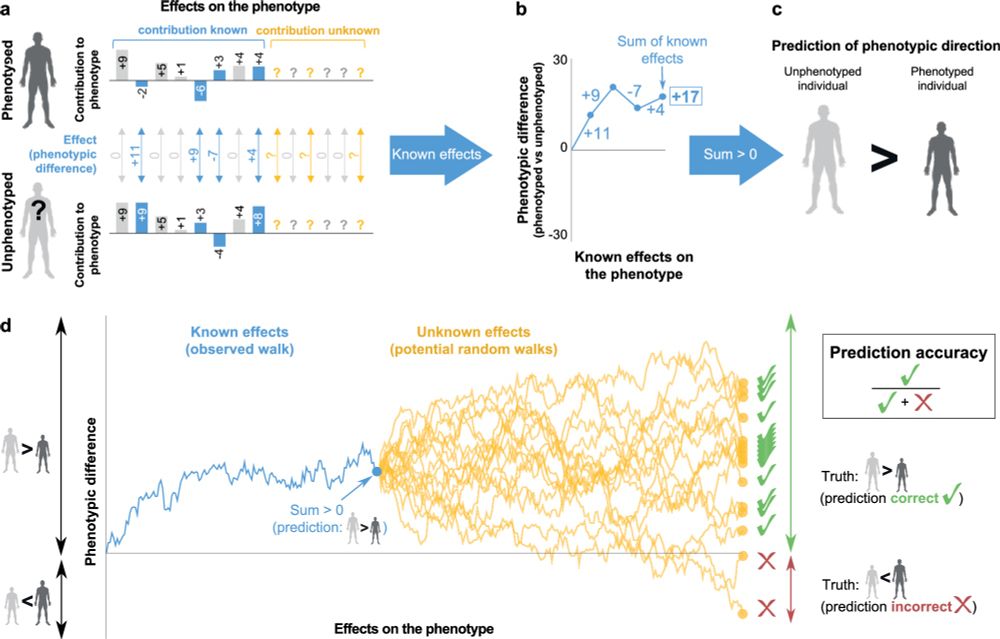
Given two genomes, can you tell who’s taller or more prone to a disease? How confident can you be? A fresh take on phenotypic inference, now out:
rdcu.be/exW4f
See thread🧵👇:
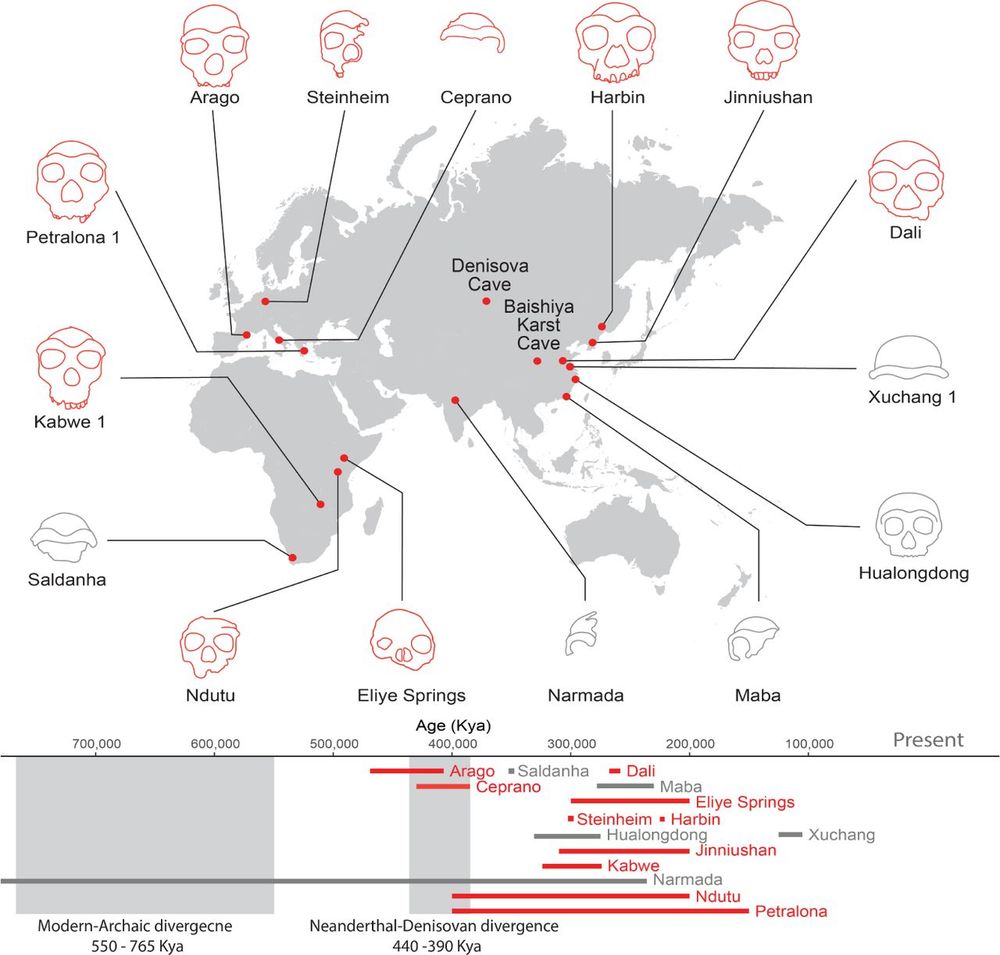
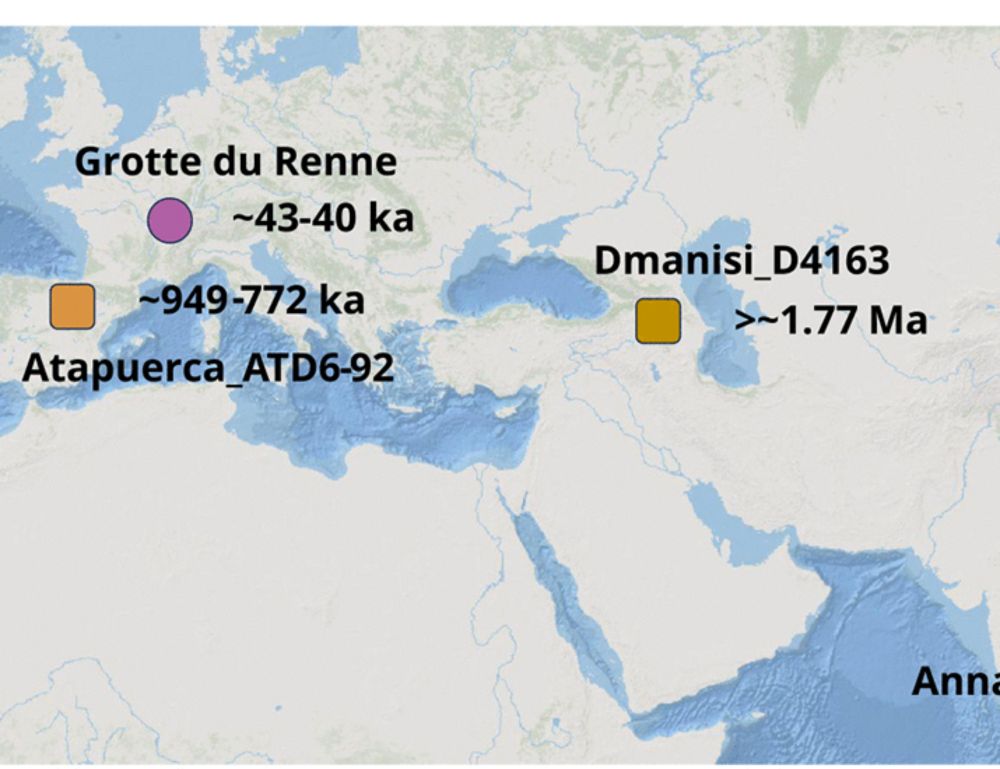
For additional specimens we think likely belong to Denisovans, see Nadav Mishol's beautiful preprint, in collaboration with
@lirancarmel.bsky.social www.biorxiv.org/content/10.1...
Although a Denisovan pinky bone has been discovered, testing our pinky-related prediction also requires the proximal and intermediate phalanges. Once those are found, we’ll also be able to test whether Denisovan phalanges exhibit the predicted tapering from proximal to distal
20.06.2025 10:33 — 👍 0 🔁 0 💬 1 📌 0Our approach was based on gene regulatory phenotyping, which predicts the direction, rather than magnitude, of phenotypic difference - a much more attainable goal. If you're interested in why and how this approach works, see: www.biorxiv.org/content/10.1...
20.06.2025 10:33 — 👍 1 🔁 0 💬 1 📌 0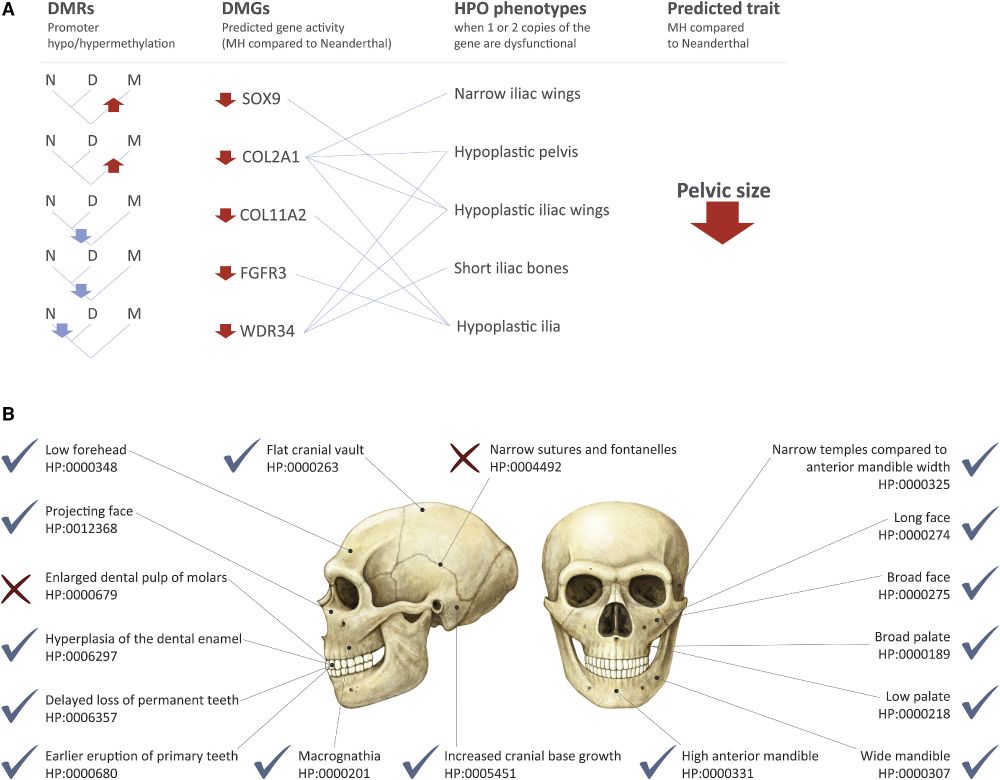
Interestingly, our Denisovan trait prediction accuracy now stands at 89% - matching the accuracy we estimated using Neanderthal and chimpanzee data. Always nice when the model does what it says on the tin.
20.06.2025 10:33 — 👍 0 🔁 0 💬 1 📌 0Overall 25/28 of our predictions have been confirmed. Not too shabby for profiling an extinct species from DNA methylation maps!
20.06.2025 10:33 — 👍 0 🔁 0 💬 1 📌 0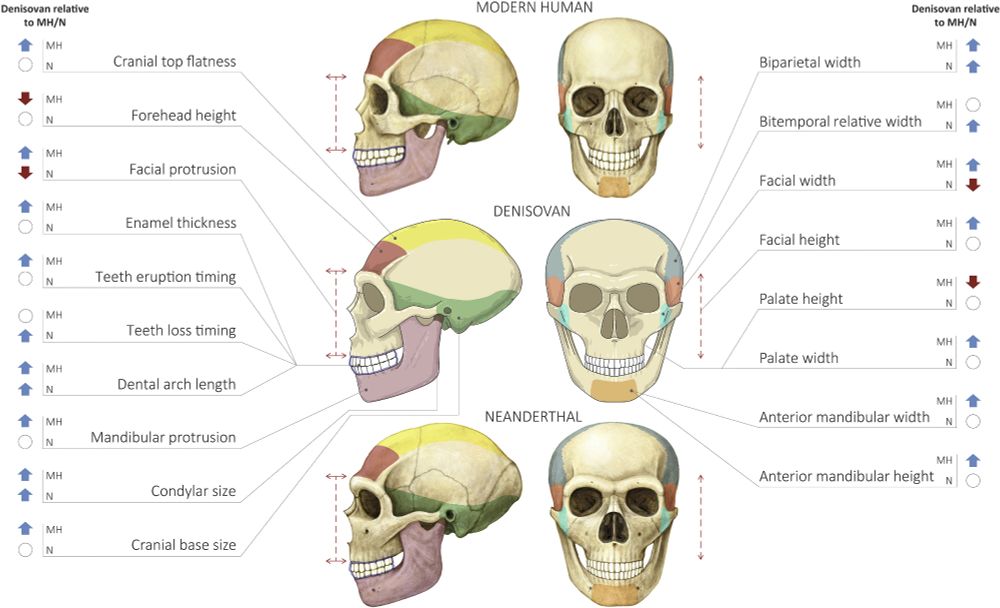
6 years ago, we predicted what Denisovans probably looked like. Since then, every new Denisovan specimen💀 - Harbin, Penghu, Xiahe - matched it beautifully.
☑️ Xiahe: 4/4 features match the profile
☑️ Penghu: 5/6
☑️ and now: Harbin with 16/18!
See 🧵
pubmed.ncbi.nlm.nih.gov/31539495/
Correct links here:
www.science.org/doi/10.1126/...
and
www.cell.com/cell/fulltex...
And the first genetic evidence supporting this was posted last year: www.biorxiv.org/content/10.1...
18.06.2025 18:42 — 👍 1 🔁 1 💬 0 📌 0First genetic evidence that Harbin was likely a Denisovan was here: www.biorxiv.org/content/10.1...
18.06.2025 18:26 — 👍 2 🔁 0 💬 1 📌 0We showed it first: www.biorxiv.org/content/10.1...
18.06.2025 18:18 — 👍 8 🔁 3 💬 0 📌 1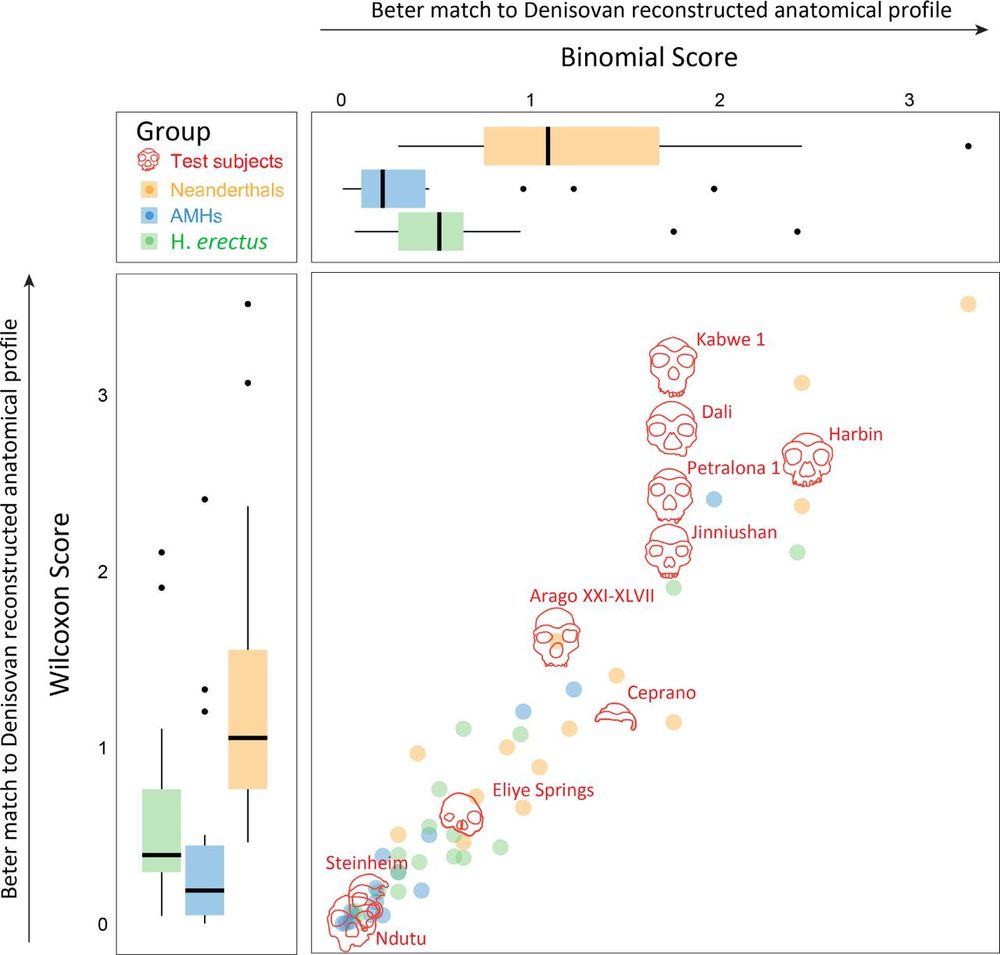
Last year, we posted a preprint providing the first genetic evidence that the Harbin skull likely belonged to a Denisovan: www.biorxiv.org/content/10.1... Today, DNA evidence provided the final proof: www-sciencedirect-com.ezproxy.weizmann.ac.il/science/arti...
18.06.2025 18:17 — 👍 2 🔁 0 💬 1 📌 0It is! And yes, we’re going to have brain-related data soon and I’m super curious to see if the variants we report played a role in human brain evo too!
07.05.2025 20:22 — 👍 1 🔁 0 💬 0 📌 0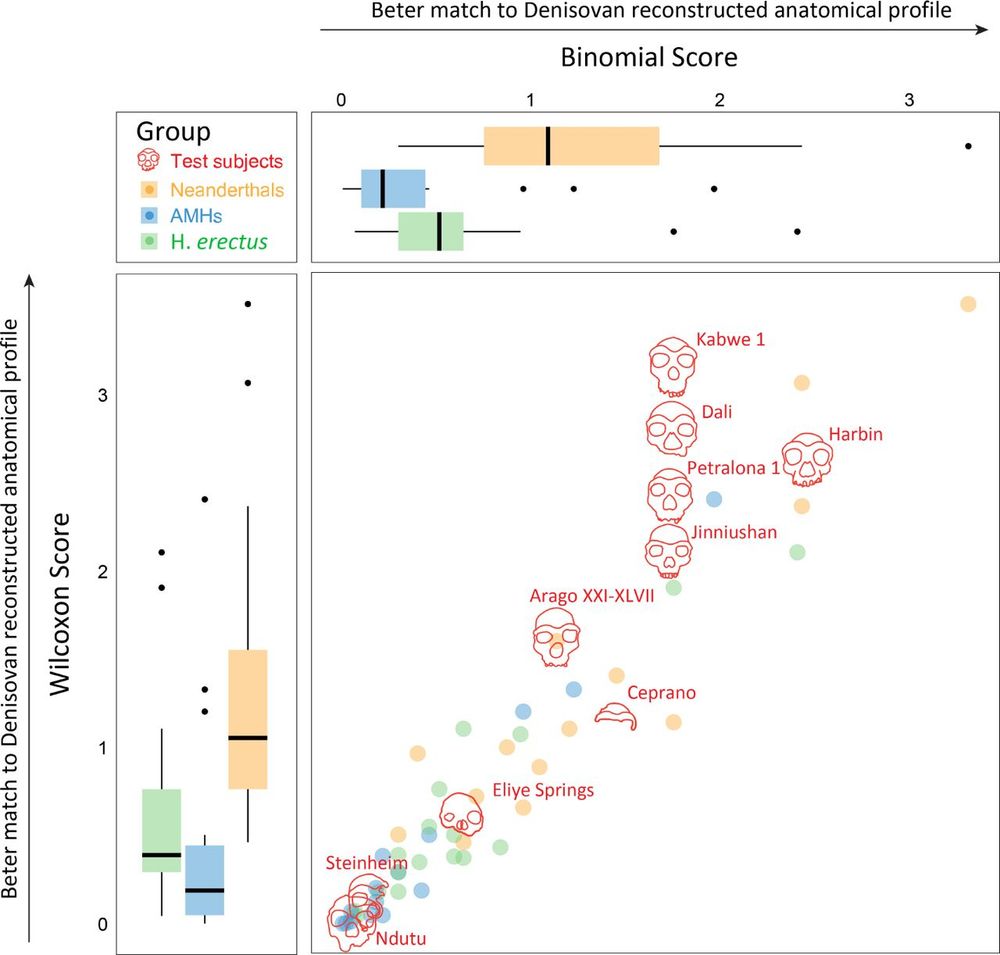
Find out about other potential Denisovan specimens here, in a beautiful work led by Nadav Mishol: www.biorxiv.org/content/10.1... @lirancarmel.bsky.social
18.04.2025 16:27 — 👍 2 🔁 0 💬 0 📌 0A common mistake when evaluating the profile is normalizing features (e.g., by breadth). But since the profile includes independent predictions affecting length, height, and breadth, normalization cancels out key signals. Absolute values are the way to go here, and they align with our predictions.
18.04.2025 16:27 — 👍 0 🔁 0 💬 1 📌 0Condyle is shorter but much broader, giving it a larger area than in humans or Neanderthals (see. Ishwarkumar et al. 2016). Symphyseal height is 31 mm in Penghu vs. 32 mm in humans.
18.04.2025 16:27 — 👍 0 🔁 0 💬 1 📌 0Technical details: anterior width (I1–I2 alveolar length) is 13 mm in Penghu vs. 11 mm median in modern humans. Prognathism (superior mandibular length) is 104 mm vs. 101 mm. Dental arch wasn’t directly measured, but Penghu is broader with a higher length/breadth ratio, indicating greater length.
18.04.2025 16:27 — 👍 0 🔁 0 💬 1 📌 0Compared to Neanderthals, we predicted a longer dental arch and a larger condyle. Except for mandibular height, all check out!
18.04.2025 16:27 — 👍 0 🔁 0 💬 1 📌 0Just like the Xiahe jaw, most of our Denisovan predictions hold up in Penghu—5 out of 6 anatomical predictions matched! 🦴✨ Compared to modern humans, Denisovans were predicted to have a wider anterior mandible, a more prognathic and taller mandible, a longer dental arch, and a larger condyle.
18.04.2025 16:27 — 👍 0 🔁 0 💬 1 📌 0