
Really interesting quantitative genetic data/modeling work in @evolletters.bsky.social: Predicting the contribution of single trait evolution to rescuing a plant population from demographic impacts of climate change
doi.org/10.1093/evle...
@hancockzb.bsky.social
Evolutionary biologist, science writer, YouTuber. Assistant Professor at Augusta University. he/him 🏳️🌈 http://www.youtube.com/@talkpopgen

Really interesting quantitative genetic data/modeling work in @evolletters.bsky.social: Predicting the contribution of single trait evolution to rescuing a plant population from demographic impacts of climate change
doi.org/10.1093/evle...
The most important paper in evolutionary biology I'd never heard of:
1/
www.sciencedirect.com/science/arti...

Delighted that our paper about the distribution of genomic spans of clades/edges in genealogies (ARGs), and using this for detecting inversions and other SVs (and other phenomena that cause local disruption of recombination) is out in MBE academic.oup.com/mbe/article/... (1/n)
03.10.2025 09:54 — 👍 61 🔁 37 💬 4 📌 1...and if you missed it, here are 60 phylogenetically diverse brown algae genomes
www.cell.com/cell/fulltex...
NSF unexpectedly changed (at the last minute) who is eligible to apply for the Grad Research Fellowship, dropping 2nd year students. We started a petition to reverse this unfair change, Sign here:
laurenkuehne.github.io/grfpChanges/
and please spread the word!!!!

New — with @joannarifkin.bsky.social, @jotlovell.bsky.social, @spicybotrytis.bsky.social, and many more — we created seven new high-quality genomes and explored pangenomic variation in the emerging oilseed crop pennycress (Thlaspi arvense). 1/
28.09.2025 17:25 — 👍 55 🔁 23 💬 1 📌 1
Another year using @kitchensjn.bsky.social + @gcbias.bsky.social's intuitive circle plots to discuss human genetic variation in my undergrad genetics class james-kitchens.com/blog/visuali...
27.09.2025 11:40 — 👍 46 🔁 16 💬 1 📌 1
Genomic divergence across the tree of life 🧪🌐https://www.pnas.org/doi/10.1073/pnas.2319389122
27.09.2025 09:21 — 👍 78 🔁 25 💬 1 📌 1
k-mer-based diversity scales with population size proxies more than nucleotide diversity in a meta-analysis of 98 plant species.
academic.oup.com/evlett/artic...

Map of the US titled: “How big is the teaching penalty in your state?” Map shows the relative teacher pay penalty for each state, ranging from 38.5% to 10%
The US has been leaving school teachers behind for nearly 30 years.
In 20 states, teachers made 25% less than their peers.
What’s the teacher pay gap in your state?
Find out here: www.epi.org/publication/...
Genetic assimilation and accommodation shape adaptation to heat stress in a splash pool copepod https://www.biorxiv.org/content/10.1101/2025.09.09.675189v1
15.09.2025 21:33 — 👍 5 🔁 2 💬 0 📌 0Very excited to see this work in press! I think there is a reason to believe that this is a common means of stabilizing large-effect polymorphisms in general and might be an important reason for why diploidy is so common. news.stanford.edu/stories/2025...
15.09.2025 17:24 — 👍 67 🔁 29 💬 1 📌 1
Hybridization and introgression are major evolutionary processes. Since the 1940s, the prevailing view has been that they shape plants far more than animals. In our new study (www.science.org/doi/10.1126/...
), we find the opposite: animals exchange genes more, and for longer, than plants

Graph showing a positive correlation between "bias in adaptive outcome" on the vertical axis and "mutation bias" on the horizontal axis, with 14 data points representing 14 respective species, with a line of best fit equal to a slope of 0.82 on the log-scale axes. Each data point contains horizontal and vertical error bars to illustrate the uncertainty in both the mutational and adaptive data. The 95% confidence interval of the slope is shown in parentheses (0.44 to 1.24), based on 10,000 simulated data sets, where the simulated data sets are based on statistical resampling of the empirical data (i.e. "bootstrap" data sets). Regression lines based on these bootstrap data sets are shown as light gray lines. The type of bias reported here refers to the ratio of transitions versus transversions, where transitions are a DNA mutation that preserves the basic structure of the DNA base (i.e. a purine-to-purine or pyrimidine-to-pyrimidine mutation), and transversions are mutations that alter the structure of the DNA base at a particular site (i.e. replacing a purine with a pyrimidine or vice versa). In other words, this graph shows that transitions contribute more toward adaptive evolution in species where transitions arise at a higher rate.
NEW PAPER! Some kinds of genetic mutations are more likely to arise than others. Such "biases" in mutation vary between species.
Analyzing data from 14 species, we show that this variation explains species differences in the genetics of adaptation!
(1/3)
www.biorxiv.org/content/10.1...

Excited to share a new paper! "Sorting of ancestral polymorphism and its impact on morphological phylogenetics and macroevolution". Part of some work I've been doing on modelling the evolution of polymorphic traits in fossil echinoderms.
academic.oup.com/evolut/advan...

Puente-Lelievre et al. have systematically reviewed how protein structures can be used to trace evolutionary histories. This rapidly advancing field has not yet caught up with the burgeoning databases of high quality protein structure predictions provided by AlphaFold and other tools.
In a new GBE Review, @cpuentelelievre.bsky.social @proteinmechanic.bsky.social & J. Douglas give an overview on protein structural phylogenetics, how to obtain evolutionary insights from structural data, and key applications and future directions.
🔗 doi.org/10.1093/gbe/evaf139
#genome #evolution




Last lecture of summer popgen:"Evolutionary QuantGen: why (almost) everything we learned is (kinda) wrong". how selection on traits affects alleles. Architecture, stabilizing selection, popgen on gene sets, corn example & a shiny app from @mgstetter.bsky.social mgstetter.shinyapps.io/quantgensimA...
25.08.2025 02:20 — 👍 47 🔁 14 💬 1 📌 0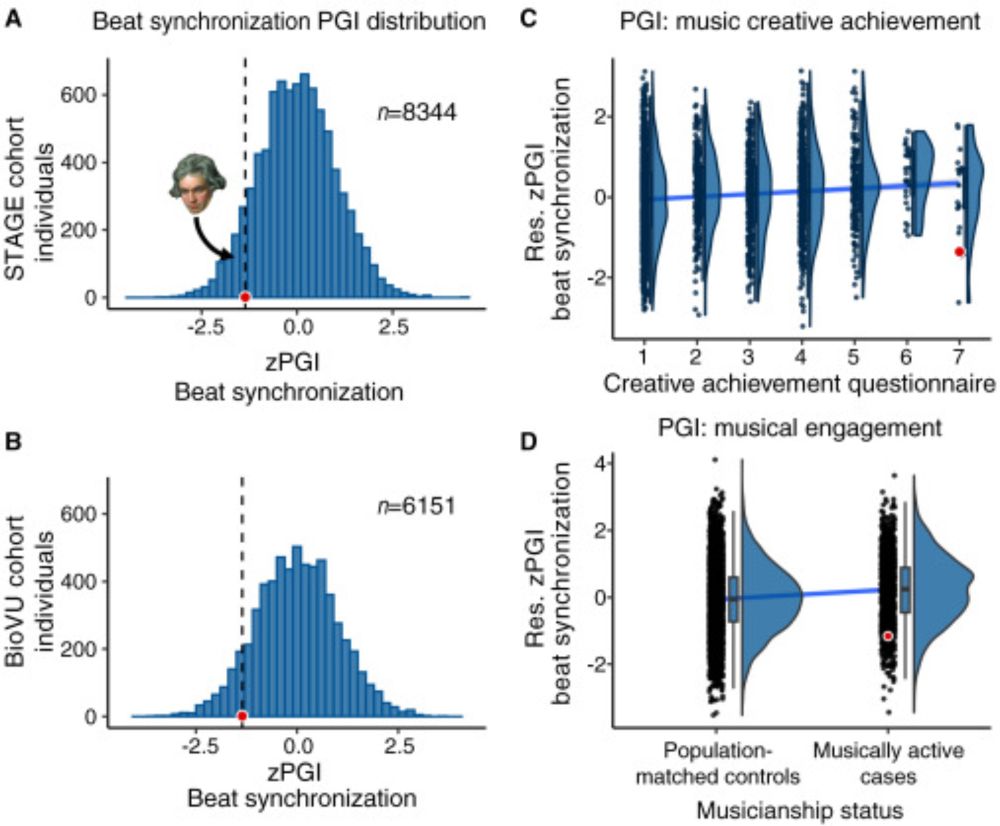
If someone you know buys into claims about "genetic optimization" of embryos using polygenic scores of cognition, just send them our 2024 paper on Beethoven & musicality. We wrote it to help communicate limits of individual-level genetic predictions & complexity of links between DNA & behaviour. 🧪👇
07.08.2025 11:09 — 👍 180 🔁 88 💬 7 📌 10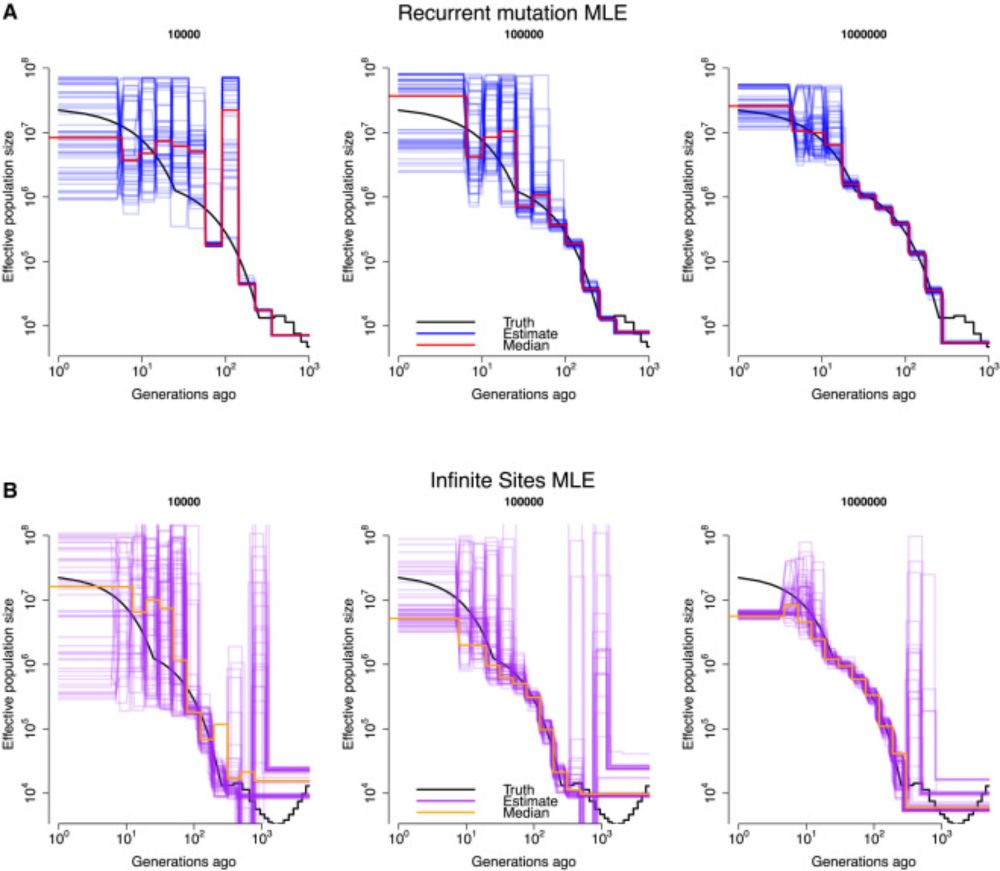
📣Online NOW!
📄Estimation of demography and mutation rates from one million haploid genomes
🧑🤝🧑 @jgschraiber.bsky.social @jeffspence.github.io @docedge.bsky.social

Our paper showing that variation in transcription factor binding sites underlies the majority of additive genetic variance for phenotypic variation in maize is finally out!
Sadly they didn't use our suggested cover image below (made by the inimitable Andi Kur).
www.nature.com/articles/s41...
SLiM 5: Eco-evolutionary simulations across multiple chromosomes and full genomes https://www.biorxiv.org/content/10.1101/2025.08.07.669155v1
11.08.2025 21:32 — 👍 58 🔁 37 💬 0 📌 1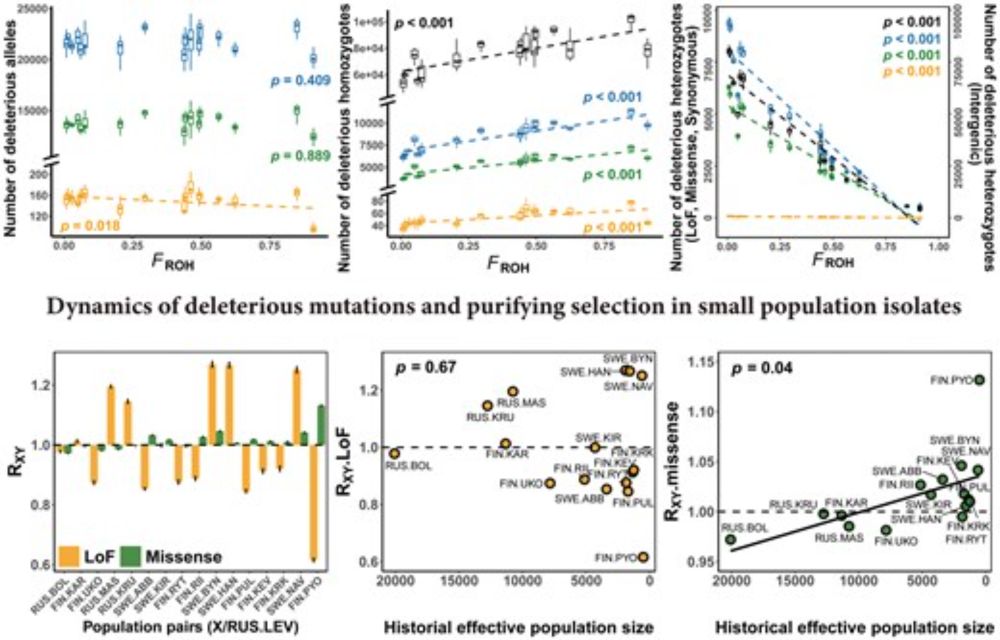
Chen et al investigate deleterious mutations and purifying selection in 17 stickleback populations, finding that inbreeding predicts realized loads of deleterious mutations, and genomic consequences of isolation in small populations are predictable.
🔗 doi.org/10.1093/molbev/msaf110
#evobio #molbio
Biologist folk (especially in evolutionary biology and/or ecology, but it don’t matter):
Can you give me your favorite examples of trade offs in biology? Organism or system don’t matter. Primary literature or reviews preferred.
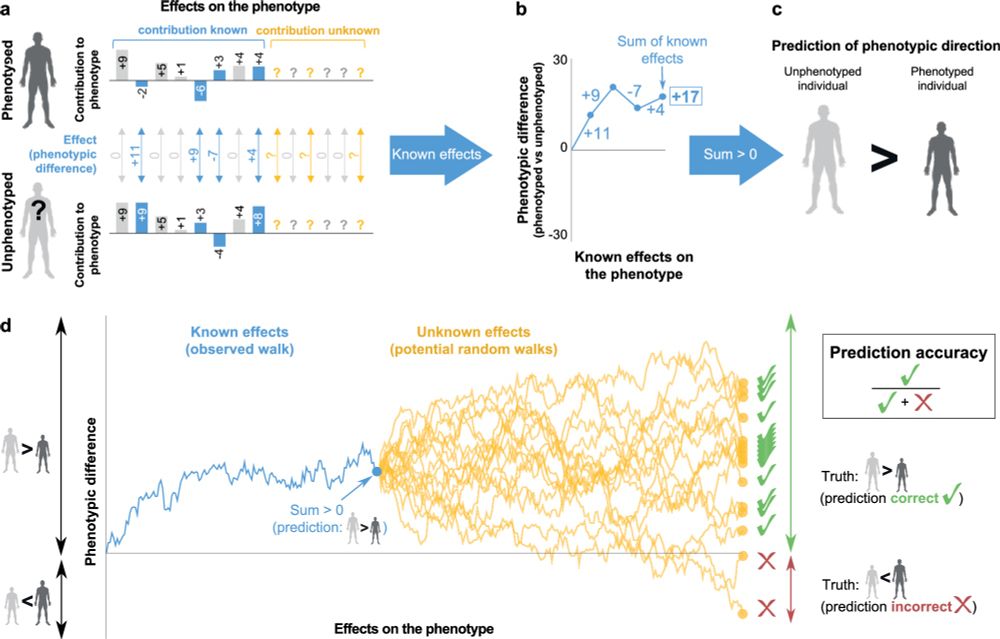
Given two genomes, can you tell who’s taller or more prone to a disease? How confident can you be? A fresh take on phenotypic inference, now out:
rdcu.be/exW4f
See thread🧵👇:
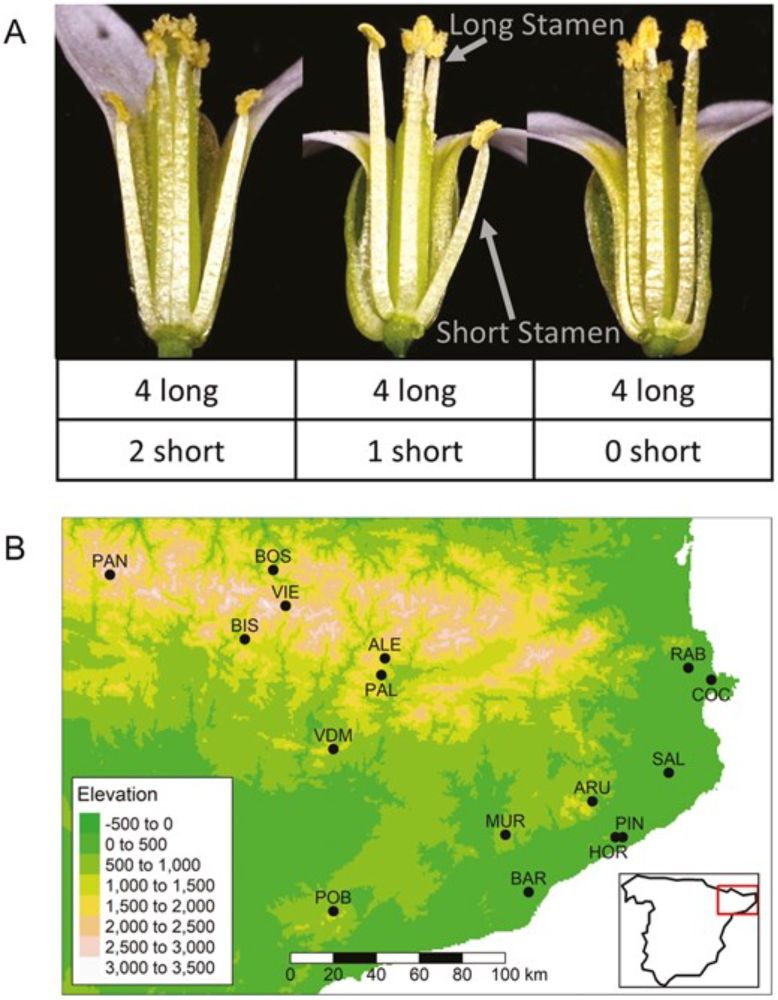
Sophie Buysse's paper on short stamen loss in Arabipsis is published in this month's Evolution issue!! academic.oup.com/evolut/artic... @journal-evo.bsky.social
21.07.2025 17:51 — 👍 28 🔁 7 💬 1 📌 2
Hopeful monsters? Great talk from @petrovadmitri.bsky.social at #SMBE2025 about dimensionality of adaptation. Here's one of the papers he mentioned (shift from pleiotropy to modularity) with @grantkinsler.bsky.social &co in @plosbiology.org: journals.plos.org/plosbiology/...
22.07.2025 03:02 — 👍 18 🔁 7 💬 0 📌 0No new genes needed to fly - just rewire what you have! 🦇🧬
Great new paper from the labs of @fany-real.bsky.social @stemundi.bsky.social @dariloops.bsky.social
doi.org/10.1038/s415...
#EvoDevo #SingleCell #BatWings
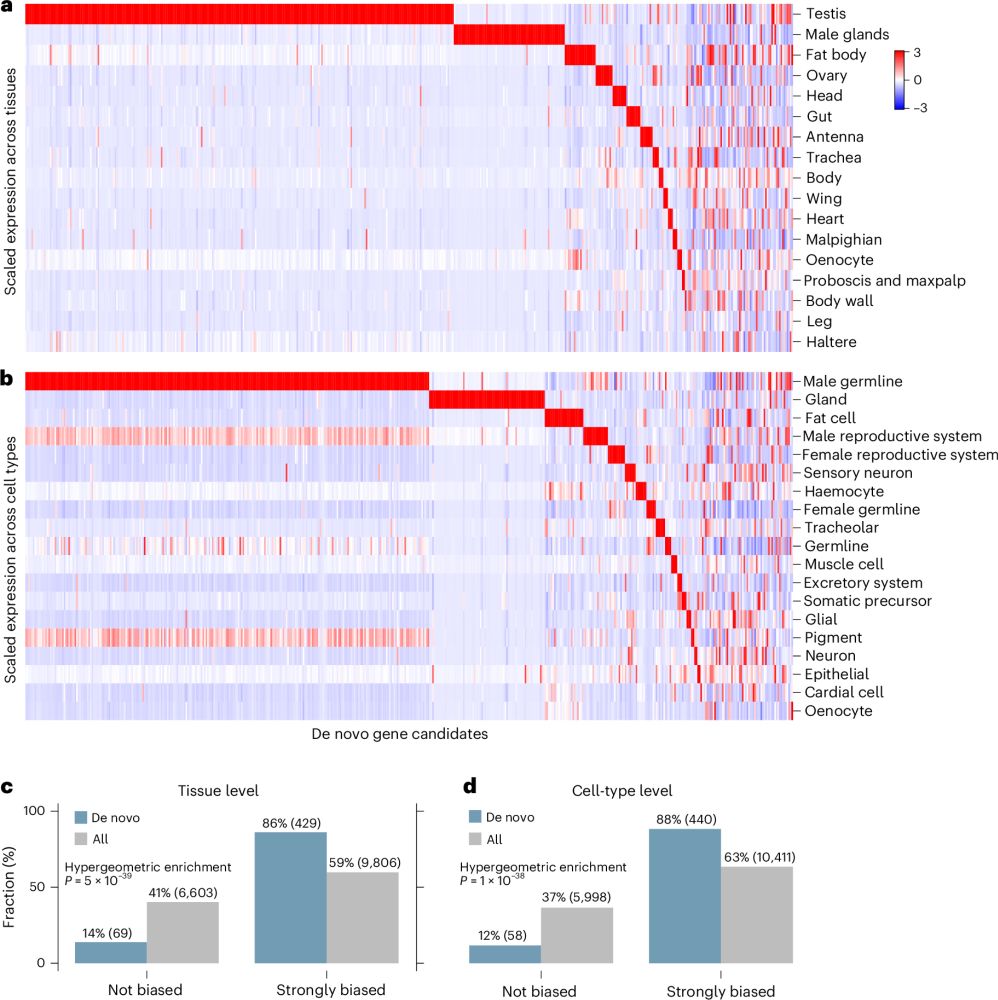
Our latest paper is out: rdcu.be/ev6Ym — one of my favorite projects. It began about 8 years ago when Nobel laureate Torsten Wiesel asked me: what transcription factors regulate new genes? I had no idea then. Now we have some answers.
15.07.2025 14:27 — 👍 111 🔁 37 💬 3 📌 3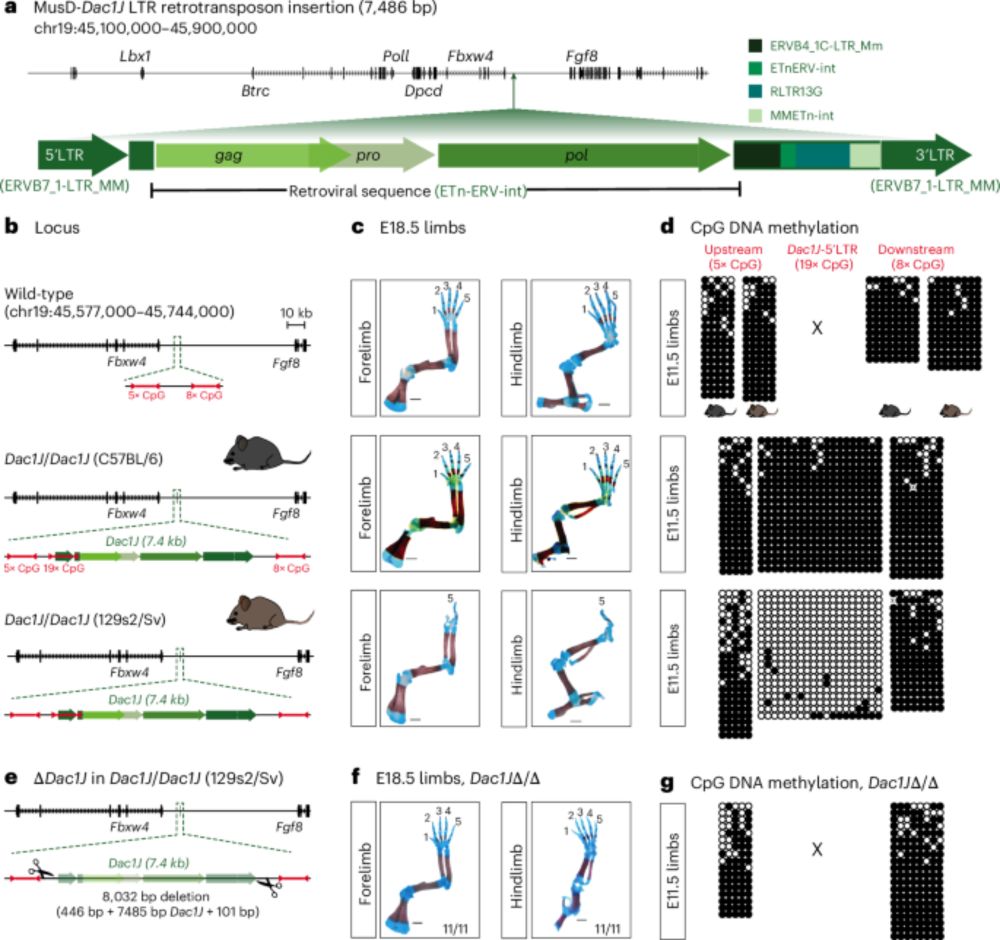
Finally out! 🥳 Our paper showing how a transposable element (TE) insertion can cause developmental phenotypes is now published @natgenet.nature.com 🧬🦠🐁
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
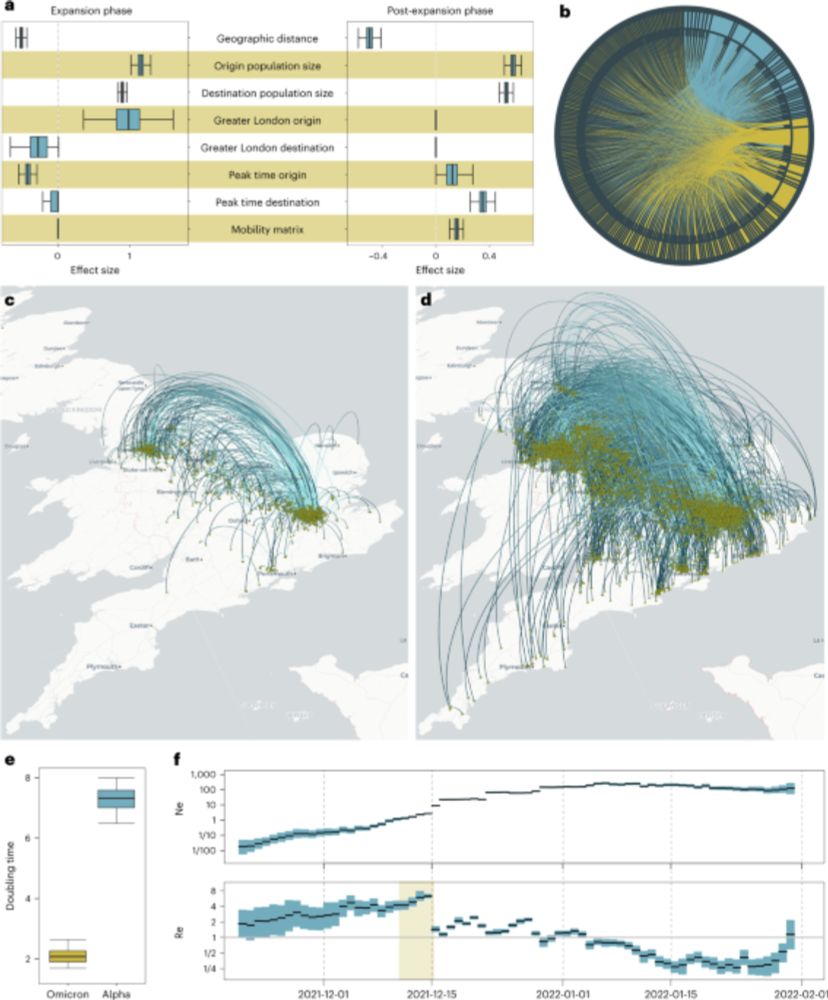
BEAST X for Bayesian phylogenetic, phylogeographic and phylodynamic inference
www.nature.com/articles/s41...