Preprint: We discovered phage proteins that sequester diverse immune signaling molecules, including cUMP, cCMP, and N7-cADPR
The first viral sponges to inhibit Pycsar and type IV Thoeris
Congrats to talented leading author Romi Hadary! Read her thread to learn more about our findings
25.08.2025 13:00 — 👍 48 🔁 15 💬 0 📌 1
New pre-print from Doudna lab members @erinedoherty.bsky.social & @jnoms.bsky.social ✨
Here we explore how recurrent adaptation of a single anti-defense protein fold enables evasion of distinct immune systems.
22.08.2025 19:36 — 👍 53 🔁 21 💬 0 📌 1

Divergent viral phosphodiesterases for immune signaling evasion
Cyclic dinucleotides (CDNs) and other short oligonucleotides play fundamental roles in immune system activation in organisms ranging from bacteria to humans. In response, viruses use phosphodiesterase...
Excited to share our new preprint co-led by @jnoms.bsky.social!
Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.
22.08.2025 19:02 — 👍 42 🔁 28 💬 2 📌 2
Fantastic results from the very talented @romihadary.bsky.social et al. on viral counter-defence sponges!! 🧽🦠
25.08.2025 13:36 — 👍 1 🔁 0 💬 0 📌 0
It was such a pleasure to work on this wonderful project with the talented @nitzantal.bsky.social !
29.07.2025 19:11 — 👍 3 🔁 0 💬 0 📌 0

END nucleases: Antiphage defense systems targeting multiple hypermodified phage genomes
Prokaryotes carry clusters of phage defense systems in defense islands that have been extensively exploited bioinformatically and experimentally for discovery of immune functions. However, little effo...
We've identified a group of nucleases that can target multiple phages with hypermodified DNA, using a sensor domain present across many domains of life. We’ve even used this as a tool to find phages with hypermodified DNA! Really thankful to all the co-authors who have been such great help!
02.04.2025 16:39 — 👍 31 🔁 7 💬 1 📌 2
Interested in phage defenses that natively block lytic phage used in therapies?
Or do you want to figure out if a phage has a modified genome?
Meet the END-nucleases, an enzyme family that can broadly restrict phages with many diverse modifications. From talented post-doc Wearn-Xin Yee!
03.04.2025 16:15 — 👍 38 🔁 18 💬 1 📌 2

A miniature CRISPR-Cas10 enzyme confers immunity by an inverse signaling pathway
Microbial and viral co-evolution has created immunity mechanisms involving oligonucleotide signaling that share mechanistic features with human anti-viral systems. In these pathways, including CBASS a...
Preprint alert! ✨ In this project that I co-led with @benadler.bsky.social, we show that a miniature CRISPR-Cas10-like enzyme, mCpol, uses a novel inverse signaling mechanism to prevent the spread of viruses that attempt immune evasion by depleting host cyclic nucleotides.
Check it out:
31.03.2025 16:27 — 👍 63 🔁 30 💬 4 📌 3
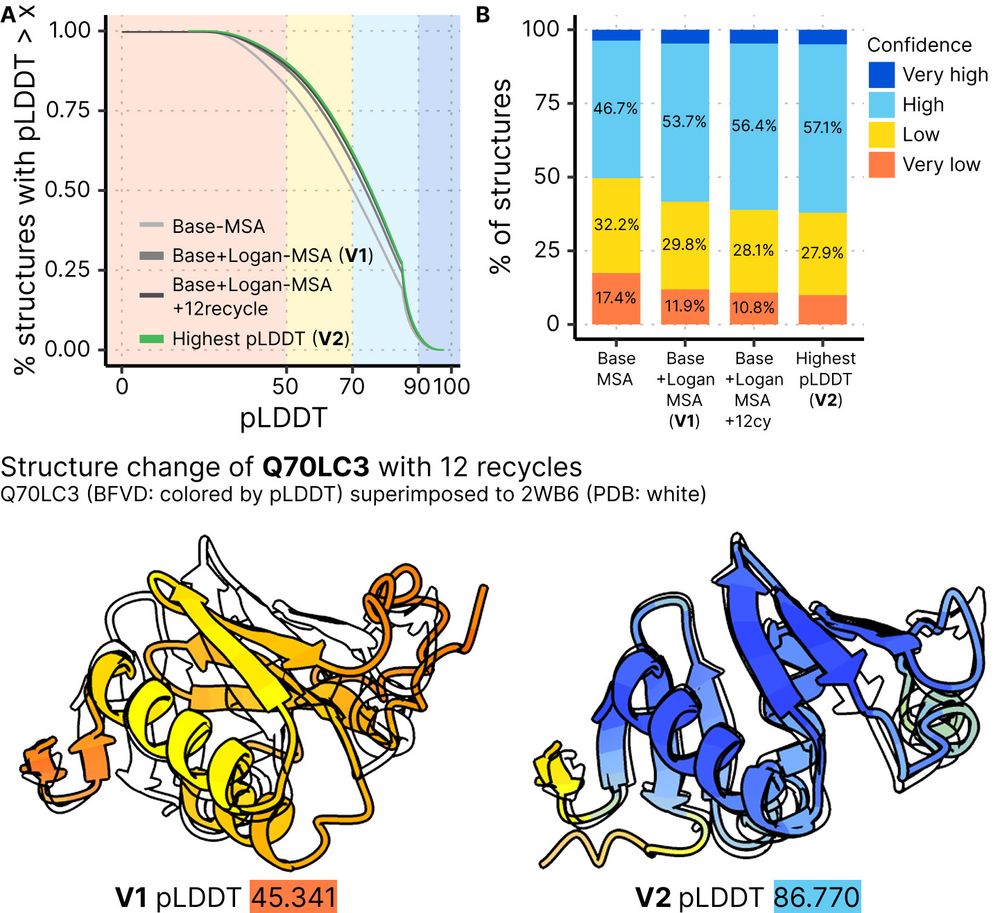
Big Fantastic Virus Database (BFVD) version 2 improves 31% of predictions through 12 ColabFold recycles. PAEs and MSAs now also available for download and in the webserver.
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
31.03.2025 05:07 — 👍 65 🔁 24 💬 2 📌 2
BFVD has been updated to version 2! Rachel improved the accuracy of viral structure predictions by running 12 recycles of ColabFold (AlphaFold2). In the latest database, over 60% of the predicted structures are now high quality. Also, we provide the PAE JSON files for each predictions.
31.03.2025 08:57 — 👍 56 🔁 16 💬 0 📌 0
A minimal CRISPR polymerase produces decoy cyclic nucleotides to detect phage anti-defense proteins https://www.biorxiv.org/content/10.1101/2025.03.28.646047v1
31.03.2025 02:17 — 👍 26 🔁 9 💬 0 📌 1
Structural basis of antiphage defense by an ATPase-associated reverse transcriptase https://www.biorxiv.org/content/10.1101/2025.03.26.645336v1
27.03.2025 03:45 — 👍 9 🔁 8 💬 0 📌 0
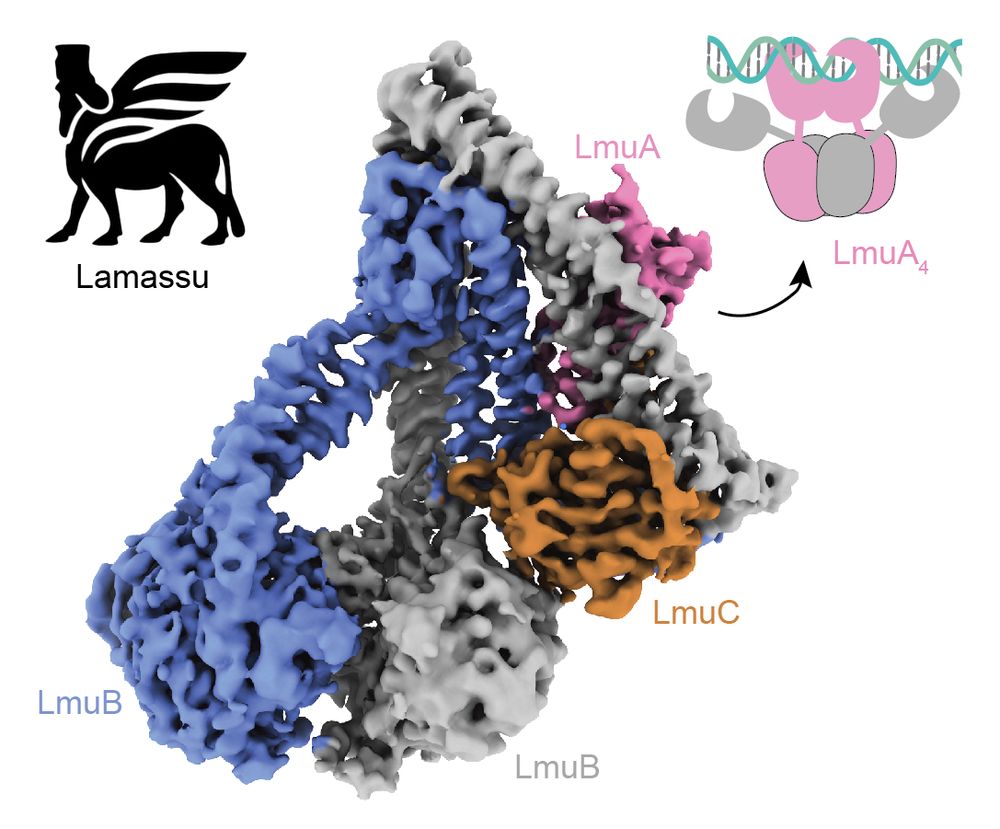
Alt : Structure of the apo Lamassu protein complex. Schematic model of the active LmuA tetramer nuclease (top right). Lamassu icon (top left).
Glad to share the work of @yli18smc.bsky.social and co on Lamassu, a bacterial defense system related to Rad50/Mre11 (RM). While RM carefully trims DNA ends for repair, Lamassu chops up the host chromosome. Our study reveals how it is regulated to minimize damage, activating only during infection.
16.03.2025 09:15 — 👍 58 🔁 30 💬 4 📌 2
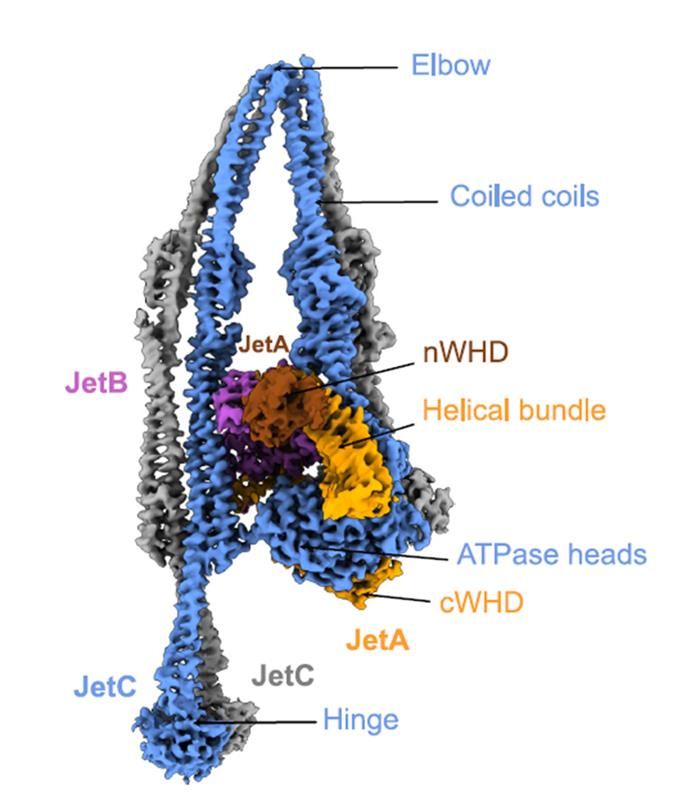
Side view of the structure of a monomer of type II Wadjet.
Happy to share the beautiful structure of a Wadjet SMC complex by @roisnehamelinf.bsky.social, with support from the DCI and @drhonsworth.bsky.social ! This is a type II Wadjet with unique characteristics including a tandem KITE subunit, distinct coiled coil architecture and a deviant hinge.
11.03.2025 09:22 — 👍 96 🔁 33 💬 1 📌 2
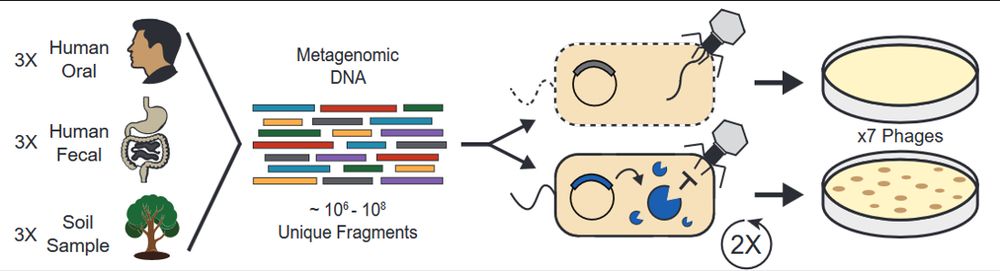
Functional Metagenomic Selections for Phage Defense
My lab’s first preprint!
We used functional metagenomics to identify phage defenses in human and soil microbiomes. We scaled these selections while maintaining accuracy, enabling us to examine 9 habitats for defense elements against 7 phages.
www.biorxiv.org/content/10.1...
1/10
03.03.2025 12:11 — 👍 152 🔁 49 💬 6 📌 2
we recently found some really neat RNA-guided DNA-cutting systems in phages. Despite remarkable similarities to CRISPR systems, including encoding guide RNAs in arrays, they appear entirely evolutionarily distinct (but definitely related to snoRNAs 🤓)
We decided to call them TIGR-Tas systems 🐯
01.03.2025 00:18 — 👍 209 🔁 79 💬 2 📌 4
Pseudomonads coordinate innate defense against viruses and bacteria with a single regulatory system https://www.biorxiv.org/content/10.1101/2025.02.26.640152v1
28.02.2025 03:19 — 👍 7 🔁 3 💬 0 📌 1
A prophage-encoded anti-phage defense system that prevents phage DNA packaging by targeting the terminase complex https://www.biorxiv.org/content/10.1101/2025.02.27.640495v1
28.02.2025 03:20 — 👍 12 🔁 5 💬 0 📌 1
Legitmately thrilled to share our latest work, in which @fernpizza.bsky.social solved an experimental challenge in plasmid biology as old as the field: measuring how plasmids compete and evolve within individual cells!
21.02.2025 20:42 — 👍 218 🔁 87 💬 5 📌 11
Principal Investigator at @aithyra.bsky.social | Structural bioinformatics, virology, and innate immunity | jasonnomburg.com
PhD Candidate in the Kranzusch Lab at Harvard Medical School / studying mechanisms of viral immune evasion
Harvard Medical School, Dana Farber Cancer Institute
https://kranzuschlab.med.harvard.edu
PhD student @Soreklab.bsky.social @WeizmannScience
Studying phage-bacteria arms race🦠🛡️
Post-doc @soreklab.bsky.social on bacterial immune system. Bacteria stick together, why can't we?
Principal Investigator | Microbial Molecular Genomics Lab | https://www.rousset-lab.com |International Center for Infectiology Research (CIRI) in Lyon 🇫🇷 Studying the molecular basis of phage-bacteria interactions.
Postdoc at Bintulab.com
Viruses, chromatin regulation and RNA
Microbiologist and phage enthusiast. Researcher at UCH CEU University.
Assistant Professor of Microbiology at UVA.
My lab studies bacterial immune systems.
All opinions are my own.
taklaboratory.org
Phages! Microbiota! MGEs! All about the little critters living around and in us :)
PhD Student at the Penadés lab, Imperial College London
Compartmentalizing your metabolism, and y’know, CRISPR stuff. Account managed by grad students and postdocs 🤙
@ucberkeleyofficial.bsky.social @innovativegenomics.bsky.social @hhmi.bsky.social
savagelab.org
Scientist exploring microbes and their interactions with each other and their hosts | Nature enthusiast, hiking, (fly) fishing, and re-learning to ski in Utah ⛷️🎣 | Dog lover & occasional foodie | Adventurous toddler dad
Always curious, always learnin
Environmental & Molecular Microbiologist | Associate Professor @univlorraine.bsky.social, @LCPME_CNRS | Mobile Genetic Elements addict | HGT, Phenotypic Heterogeneity, AMR, Single Cell Analysis, Tn-seq
Assistant Professor @ ISTA
Using cryo-EM to understand how bacteria defend themselves
https://bravo-lab.org/
Investigating host-pathogen interactions across all domains of life 🌱🦠. Current focus is plants within @pcronald.bsky.social lab at @ucdavis.bsky.social. PhD in Joe Bondy-Denomy’s lab at UCSF. Website: https://erin.phd. Schmidt Science Fellow.
Research fellow at Imperial College London
PhD student at the Atkinson Lab, Lund University. Bioinformatician (mathematical biology, translation, protein evolution, data visualisation) 🇮🇱🇺🇦🇸🇪
https://scholar.google.com/citations?user=r3VBTyoAAAAJ&hl=en
Postdoc at @uclcancer with the
Fisher Lab | PhD at @emblebi |
Interested in modeling cell signaling in the contexts of disease and dysregulation
PhD candidate Biochemistry in the Swarts lab @WUR