
Opening of a Group Leader position in AI in genomics at Institut Pasteur @pasteur.fr , Paris research.pasteur.fr/en/call/mid-...
06.12.2025 09:48 — 👍 8 🔁 14 💬 0 📌 0@sylvainbrisse.bsky.social
professor at Institut Pasteur. Bacterial genomics, vaccine-preventable diseases, antimicrobial resistance, bioinformatics and public heath applications. International capacity building and teaching. Klebsiella diphtheria whooping cough genomic librairies

Opening of a Group Leader position in AI in genomics at Institut Pasteur @pasteur.fr , Paris research.pasteur.fr/en/call/mid-...
06.12.2025 09:48 — 👍 8 🔁 14 💬 0 📌 0📣🦠Check out our new Editor’s Spotlight! Senior MGEN editor Jesse Shapiro tells us about a recently published paper on the bystander effect on the evolution of Bordetella parapertussis, a non-target species of the pertussis vaccine #MGEN
Read the full blog here: microb.io/48zfOlJ

The potential for large outbreaks of pertussis highlights the importance of maintaining or increasing vaccine coverage in pregnancy and in infants and children.
02.12.2025 06:24 — 👍 0 🔁 0 💬 0 📌 0
Global Incidence of #Pertussis after the COVID-19 Pandemic - A large collaborative effort under the auspices of the International #Bordetella Society, led by Andrew Gorringe, from the UK Health Security Agency. jamanetwork.com/journals/jam...
02.12.2025 06:24 — 👍 2 🔁 1 💬 1 📌 0And a big thanks to our funders @gatesfoundation.bsky.social @pasteur.fr @ibeidlabex.bsky.social @wellcometrust.bsky.social @klebnet.bsky.social
30.11.2025 19:51 — 👍 1 🔁 0 💬 0 📌 0Thanks to @kjolley.bsky.social @chcrestani.bsky.social @kelwyres.bsky.social @margaretlam.bsky.social Martin Maiden @pubmlst.org @daanensen.bsky.social @katholt.bsky.social Federica Palma, Melanie Hennart and Alexis Criscuolo @giphy-ip.bsky.social and other co-authors
30.11.2025 19:51 — 👍 4 🔁 0 💬 1 📌 0The wide adoption of cgMLST LIN code strain taxonomies has the potential to result in a universal approach for standardized bacterial genotyping that could greatly enhance microbial biodiversity studies, international genomic epidemiology and infectious disease surveillance
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
Data confidentiality is an important barrier to rapid response to outbreaks. As LIN codes are sufficient to define strain identity across countries, they provide a solution for cross-border collaborative genomic surveillance investigations, without the need to share genomic sequences themselves
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
The recognition of sub-populations associated with distinct phenotypes is an important “raison d’être” of taxonomies. cgMLST LIN codes strain taxonomies will advance our understanding of the links between genotypes and clinical phenotypes, vaccine coverage and antimicrobial resistance
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
LIN codes provide clear definitions of sublineages, can help track dissemination at fine genetic scales within sublineages, and can subdivide isolates from single long-term outbreaks
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
To make the LIN code taxonomy broadly accessible, its components (alleles, profiles, cgSTs, LIN codes and nicknames) can be extracted using an application programming interface, and used with external tools and analysis platforms, such as @pathogenwatch.bsky.social
30.11.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
A taxonomic system needs to be created and updated in a coordinated manner. The cgMLST LIN code strain taxonomy is implemented in @kjolley.bsky.social 's BIGSdb, and is deployed at two main sites, PubMLST (at Oxford University) and BIGSdb-Pasteur (at Institut Pasteur, Paris)
30.11.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
LIN code diversity within a bacterial species can be summarized using circular packing plots, which illustrate the diversity of populations at each classification level. These representations also enable the identification of the most epidemiologically represented populations
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
LIN codes can be nicknamed with simple denominations, to enable easy communication. Nicknames can be assigned to LIN code prefixes based on prior denominations. For example, Klebsiella pneumoniae sublineages (SL) and clonal groups (CG) were nicknamed using the popular MLST identifiers
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
Core genome multilocus sequence typing (cgMLST) enables standardized high-resolution strain genotyping. Life Identification Number (LIN) codes are series of numeric codes that reflect genomic similarity between organisms, from left to right down to strain identity.
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0« cgMLST LIN codes » These strange acronyms hide an important advance in the standardization of bacterial strains taxonomy, enabling harmonized global surveillance of bacterial pathogens and more efficient multi-country outbreak investigations
30.11.2025 19:51 — 👍 1 🔁 0 💬 1 📌 0
Huge preprint if you are interested in bacterial strain taxonomy! The why and how of cgMLST LIN codes: An extensively revised and expanded version doi.org/10.1101/2024... I will summarize it for you in this thread 👇
30.11.2025 19:51 — 👍 45 🔁 27 💬 2 📌 1
The next five videos from our #IMMEM XIV poster presentations are available on our YouTube channel and cover topics like #plasmids, #genomics, #sequencing and more. Stay tuned for more videos every Wednesday!
https://youtube.com/playlist?list=PLBG53xT7PX7v-C3XcKHnKeVP1NxsrLqBU&si=tpFqDe_obR6LZLtx
Great work by @armovse.bsky.social with help from @josefdelgadoblas.bsky.social and colleagues at @institutpasteur.bsky.social and @flebreton.bsky.social ‘s group
26.11.2025 14:27 — 👍 1 🔁 0 💬 0 📌 0
We show nearly equal discrimination when compared to whole-genome single nucleotide polymorphisms analysis. The cumulative cgMLST strategy combines broad phylogenetic applicability and nearly complete genotyping resolution, expanding the utility of this harmonized approach for genomic epidemiology.
26.11.2025 14:27 — 👍 1 🔁 0 💬 1 📌 0
Compared to the KpSC scheme used alone, pairwise allelic distances among isolates increased on average 5.6-fold using the Kpn scheme. Schemes are easy to combine using BIGSdb’s interface or APIs developed by @kjolley.bsky.social
26.11.2025 14:27 — 👍 0 🔁 0 💬 1 📌 0As a use case, we created non-redundant cgMLST schemes for the individual species K. pneumoniae sensu stricto (Kpn-cgMLST scheme), and its multidrug resistant sublineages (SLs) SL147 and SL307.
26.11.2025 14:27 — 👍 0 🔁 0 💬 1 📌 0
We illustrate this approach using the Klebsiella pneumoniae species complex (KpSC). I have been asked for ages, «When will you develop a more resolutive scheme ?». Here is it ! … with a systematic approach applicable to all broadly diverse bacterial species, and all sublineages thereof.
26.11.2025 14:27 — 👍 0 🔁 0 💬 1 📌 0
To overcome this trade-off, we conceptualizaed and developed the cumulative cgMLST approach, where sets of core loci conserved within nested phylogenetic entities are added.
26.11.2025 14:27 — 👍 0 🔁 0 💬 1 📌 0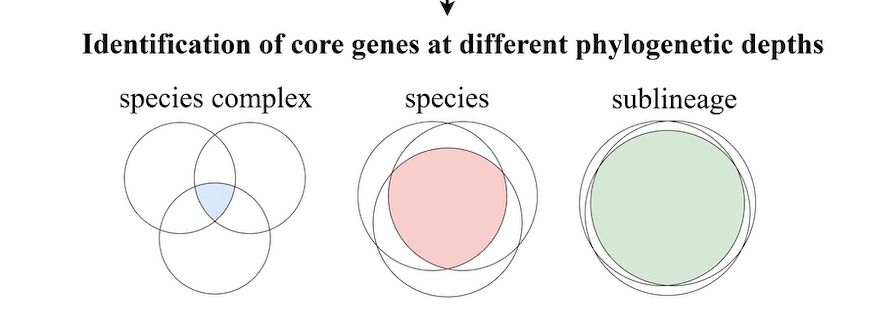
Core genome multilocus sequence typing (cgMLST) is a powerful method for bacterial strain genotyping. However, the size of the core genome decreases as the phylogenetic breadth of the target group increases, reducing discriminatory power. We call this the discrimination/applicability tradeoff
26.11.2025 14:27 — 👍 0 🔁 0 💬 1 📌 0
Cumulative cgMLST : Our novel preprint is out @biorxiv doi.org/10.1101/2025...
26.11.2025 14:27 — 👍 11 🔁 4 💬 1 📌 1
Great video on the impact of the magic bullet developed by @dndi.org against human african trypanosomiasis youtu.be/GcIx32MDdkU?...
26.11.2025 08:37 — 👍 2 🔁 0 💬 0 📌 0The @klebnet.bsky.social team are pleased to share slides from our “Klebsiella pneumoniae Genomic Epidemiology & Antimicrobial Resistance” lecture series!
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...
Great collaboration between @institutpasteur (Thanks to Valerie Bouchez and others in my group), @vallhebron.com (team of Juanjo Gonzalez) and #CDC (Mike Weigand @mikeyweigand), and @anisabelBento
16.11.2025 18:01 — 👍 0 🔁 0 💬 0 📌 0
The most recent common ancestor of all sequenced Bpp isolates was estimated around the year 1877, making it one of the youngest human pathogens. We retraced the adaptation of #parapertussis to humans via key mutations in the master regulator bvgA-fhaB intergenic region around year 1900.
16.11.2025 18:01 — 👍 0 🔁 0 💬 1 📌 0