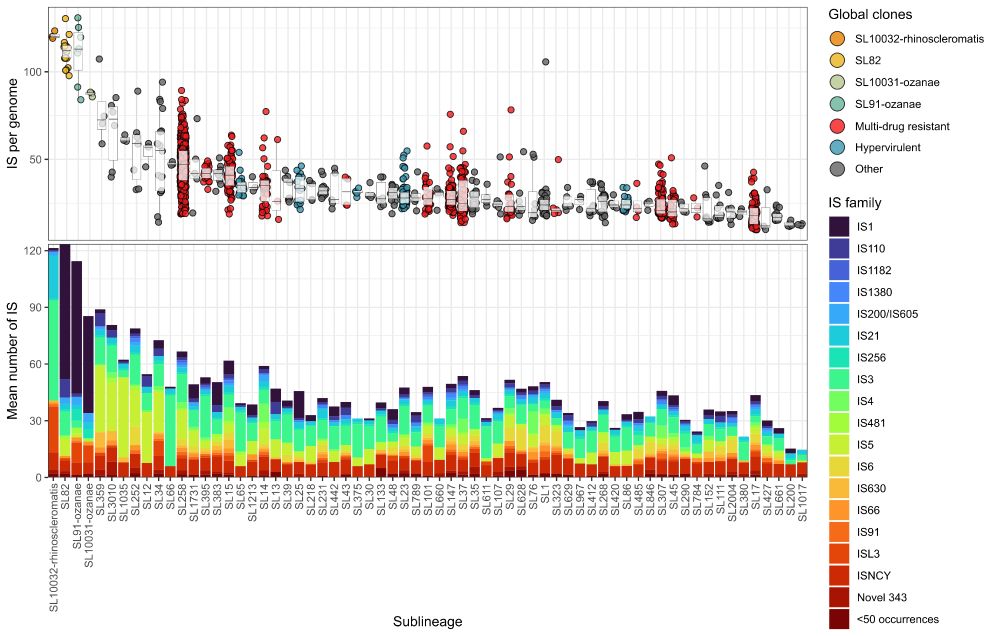
Graph showing IS loads by Klebsiella sub-lineage
New preprint where we analysed IS loads in 🦠 #Klebsiella pneumoniae lineages and found they're inversely associated with nutrient usage. We propose an insertional tolerance model, whereby IS can only insert into metabolically-tolerable sites.
#IDSky #MicroSky 🧪
www.biorxiv.org/content/10.1...
29.07.2025 02:29 — 👍 18 🔁 8 💬 1 📌 0
These findings show that Nanopore data is now accurate enough, practical and ready for further integration into clinical settings. Massive thankyou to @nenadmacesic.bsky.social, @yekwah.bsky.social, @katholt.bsky.social and all others who contributed to this project! /5
30.07.2025 02:14 — 👍 5 🔁 1 💬 0 📌 0
We provide specific recommendations for sequencing depth, basecalling model and analysis methods depending on the resources available and clinical context of the user. Analysis time is negligible, the biggest time savings come from choosing your basecalling model and depth requirements /4
30.07.2025 02:13 — 👍 5 🔁 0 💬 1 📌 0
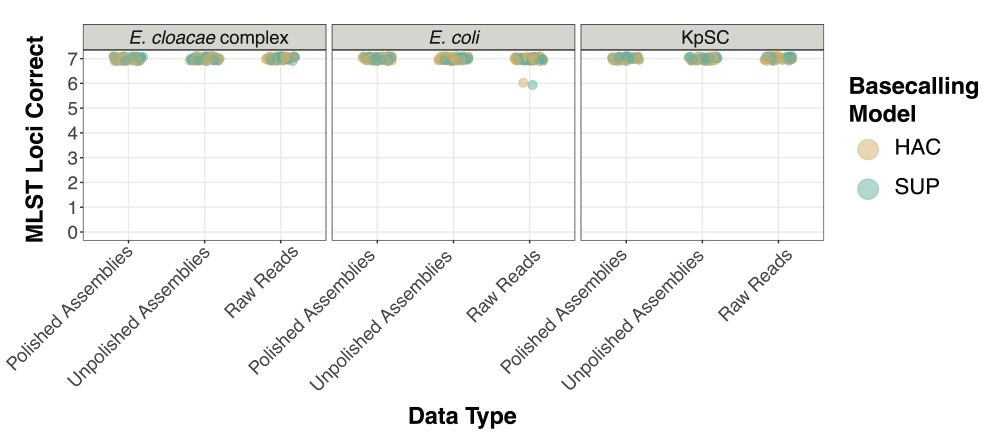
We found modern ONT data (R10.4.1, Dorado SUP) to generate perfect MLST profiles and AMR allelic variant profiles, ~99.5% cgMLST profiles and cgSNP profiles highly concordant with traditional Illumina methods /3
30.07.2025 02:13 — 👍 4 🔁 0 💬 1 📌 0
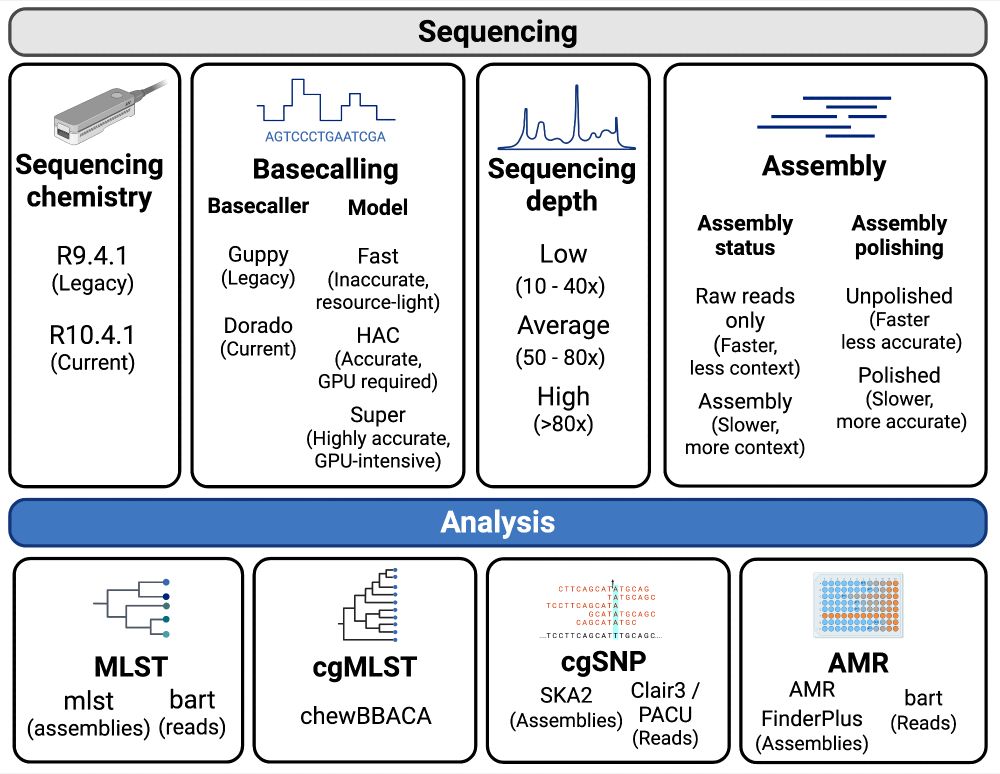
Nanopore data has come a long way the last few years, and offers users a huge amount of flexibility in how they want to generate data. We took all combinations of sequencing chemistry, basecalling, depth & assembly status from 3 Gram-negative species groups and compared to hybrid gold standards /2
30.07.2025 02:11 — 👍 3 🔁 0 💬 1 📌 0
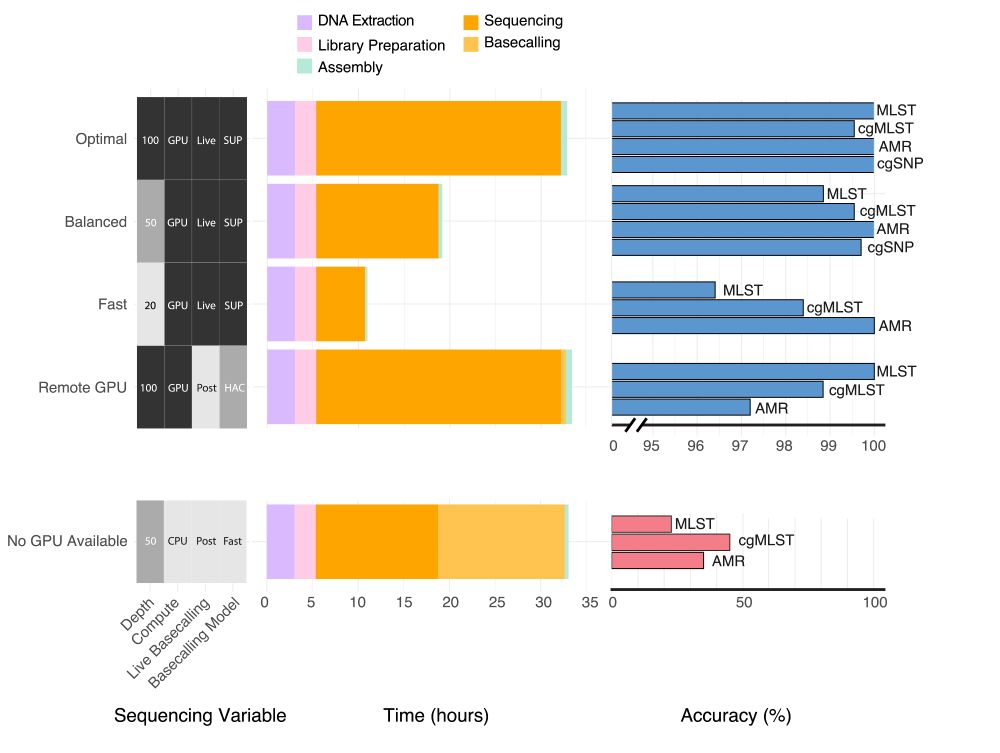
Pleased to say that our preprint benchmarking Nanopore data for MLST, cgMLST, cgSNP & AMR typing from bacterial isolates is out! TL;DR you can get almost perfect results from 50x depth using live SUP basecalling with a GPU in under 20 hours #microsky#IDsky 🦠🧬🖥️ /1
www.medrxiv.org/content/10.1...
30.07.2025 02:10 — 👍 41 🔁 27 💬 3 📌 2
Computational Biologist / Bioinformatician
at Doherty Institute and Unimelb
https://scholar.google.com.au/citations?user=CV66a6sAAAAJ&hl=en
Psyduck is my spirit animal.
Bioinformatics geek 🤓 crafting Rust-y tools 🦀 for microbial genomes 🦠 🧬.
Trying to master Dad mode 👨🍼
See what I'm up to here: https://github.com/mbhall88
Bioinformatician interested in infection and immunity. Direct from sample sequencing for diagnostics. MMM group, NDM, Oxford University
bacterial genomics・antibiotic resistance https://klebnet.org
Antimicrobial Resistance | Sustainable Solutions.
Combining Engineering, Social, Biological, Evolutionary, Chemical and Clinical solutions to AMR
ID physician in Melbourne, Australia.
professor at Institut Pasteur. Bacterial genomics, vaccine-preventable diseases, antimicrobial resistance, bioinformatics and public heath applications. International capacity building and teaching. Klebsiella diphtheria whooping cough genomic librairies
Group leader at Peter Mac & the Doherty Institute in Melbourne, Australia
microbial genomics, statistics, machine learning
Scientist… pathogen genomics & antimicrobial resistance, computational biology & infectious disease epidemiology.
Co-Director LSHTM AMR Centre @lshtmamrcentre.bsky.social
holtlab.net | klebnet.org | typhoidgenomics.org | amr.lshtm.ac.uk
Research Fellow at Monash University - microbial genomics, Klebsiella, Acinetobacter, plasmids
Lead Research Software Engineer AMRnet at @lshtm.bsky.social. Scientist from Brazilian Amazon. Advocate #amazonia #cloud #AI #AMR #onehealth #ESG.
Bioinformatician at Stavanger University Hospital & PhD fellow at University of Bergen looking at Klebsiella genomics (and sometimes other bugs).
Computational microbiologist; genomics, metabolic modelling, Klebsiella, capsules, population biology. Toddler mum and wannabe pilates enthusiast.
Postdoc with MDU @dohertyinstitute @unimelb
Interested in bacterial genomics and bioinformatics, particularly re: AMR transmission in hospitals.
Views my own. She/her.
Professor of Algorithmic and Microbial Genomics at the University of Bath (UK). Pangenomes, drug resistance (esp TB), data structures for DNA search, plasmid evolution, global microbial surveillance. Open Data, reproducibility
Scientist, using #genomics to understand #evolution, diversity, & pathogenesis. Welsh expat living in New Zealand. Formerly The University of Queensland
Infectious Disease Epidemiologist - Nontuberculous mycobacteria: Research Fellow at the University of Queensland | Microbial Genomics | Also interested in evolution, pathogenesis and #AMR of clinically relevant bacterial pathogens | She/her | Cat lover
Bioinformatician at the Centre for Pathogen Genomics at the University of Melbourne
Genomic epidemiologist at the Centre for Genomic Pathogen Surveillance at Oxford. How can microbial genomics improve health in high infectious disease settings?



