Huge thanks to everyone involved, especially my co-first authors, @ceesdekker.bsky.social , and our great collaborators at Jan-Michael Peters' lab @impvienna.bsky.social
12.12.2025 07:33 — 👍 0 🔁 0 💬 0 📌 0
CTCF doesn't just have one stop signal—it has two!The YDF & KTYQR motifs stall the cohesin loop extruder in different ways: one is a roadblock, the other forces one-way traffic.
12.12.2025 07:33 — 👍 0 🔁 0 💬 1 📌 0

🚨New paper out! In our @MolCellNews paper, we solve a piece of the puzzle of how our genome folds. We found the two specific molecular "brakes" in the CTCF protein that stop DNA loop extrusion. One of the final pieces of my PhD!
tinyurl.com/29uuc5by
12.12.2025 07:33 — 👍 21 🔁 10 💬 1 📌 2
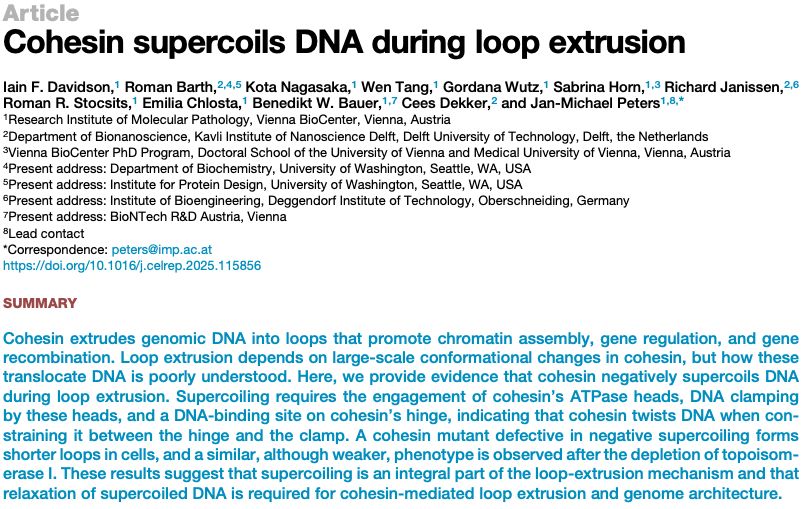
Latest #CDlab paper online now:
Cohesin supercoils DNA during loop extrusion
www.cell.com/cell-reports...
This extensive study was led by Jan Michael Peters at Vienna; our lab contributed mostly modeling of plasmid supercoiling (@romanbarth.bsky.social) and single-molecule data (Richard Janissen).
16.06.2025 12:01 — 👍 25 🔁 8 💬 1 📌 0
Congratulations to the 2025 Schmidt Science Fellows - exciting that you'll be joining the community!!
03.04.2025 15:01 — 👍 2 🔁 0 💬 0 📌 0

We have 3 openings for ambitious postdocs or PhD students in our #CDlab - for exciting single-molecule biophysics research on nuclear pores, peroxisomes, or archaeal divisomes.
Check it out and apply: ceesdekkerlab.nl/come-join-us/
RT=nice!
13.03.2025 16:37 — 👍 32 🔁 38 💬 0 📌 1

Huge congrats to our #CDlab PhD student @romanbarth.bsky.social for winning the prestigious 2025 Weintraub Award for his PhD thesis work on SMC motor proteins!
www.fredhutch.org/en/news/rele...
Great honor and well deserved Roman!
04.03.2025 19:37 — 👍 17 🔁 3 💬 0 📌 0
Science is a collaborative adventure, and this moment really feels like a celebration of the collective effort, curiosity, and passion that drives research.
05.03.2025 02:01 — 👍 0 🔁 0 💬 1 📌 0

I'm excited to share that I've been selected as one of the 10 recipients of the prestigious Weintraub Award from @fredhutch.bsky.social this year! 🏆
I owe a massive debt of gratitude to the remarkable colleagues and mentors who have shaped my path, especially @ceesdekker.bsky.social !!
05.03.2025 02:01 — 👍 25 🔁 3 💬 2 📌 1

Check out this cool article by @newscientistnl.bsky.social on our latest work about direction-switching molecular motors racing along DNA: www.newscientist.nl/nieuws/eiwit...
04.02.2025 06:59 — 👍 4 🔁 0 💬 0 📌 0
in a PDS5 knockdown? STAG2 seems to be a liiiiittle enriched there (Fig. 4c) and AF is more confident in STAG2 than in STAG1. Yet we didn't test STAG2.
Nevertheless, in the (local) absence of PDS5, CTCF might use the KTYQYR motif to interfere with cohesin as our data suggests..
30.01.2025 05:39 — 👍 0 🔁 0 💬 0 📌 0
seems to happen also experimentally. We didn't explore PDS5 binding to KTYQR computationally or experimentally. It might be that KTYQR-PDS5 binding is stronger than KTYQR-STAG binding and this is thus the dominant mode seen in vivo. Are you aware of Hi-C and/or binding (as in your Fig. 4) data..
30.01.2025 05:39 — 👍 0 🔁 0 💬 1 📌 0
Hi @elphegenoralab.bsky.social and @eecutts.bsky.social . True, the KTYQR motif has been identified by both of your cited articles as PDS5 binding regions. We didn't test PDS5 in our assays, only cohesin-NIPBL. Seeing that AF suggested KTYQR-STAG binding was surprising to us and more so that that..
30.01.2025 05:39 — 👍 0 🔁 0 💬 1 📌 0


Latest #CDlab paper now at @biorxivpreprint.bsky.social: www.biorxiv.org/content/10.1...
In this paper, we identify two amino acid motifs on CTCF that impede cohesin-mediated DNA loop extrusion - important as CTCF and cohesin are crucial to the structure and regulation of our chromosomes.
27.01.2025 09:02 — 👍 35 🔁 5 💬 1 📌 0
We started
27.01.2025 17:34 — 👍 0 🔁 0 💬 0 📌 0
Thank you to all that contributed!! Some on bluesky: @jacotorre.bsky.social @ceesdekker.bsky.social, especially to the fantastic team at the IMP Vienna Gabriele Litos, Iain Davidson, and Jan-Michael Peters!!
27.01.2025 17:34 — 👍 3 🔁 0 💬 1 📌 0
We hope that by disentangling the multiple contributions of the CTCF N-terminus to the stalling of cohesin at CTCF sites, we are another step closer to understanding how cohesin and CTCF control and shape genomes across the tree of life 🌳
27.01.2025 17:34 — 👍 3 🔁 0 💬 1 📌 0

To summarize:
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0

KTYQR is distinct from YxF: it reduces cohesin's LE activity overall, but doesn't impact cohesin's directionality.
Together, the two motifs account pretty much for the impact of the full NTR!! We hypothesize that connecting the two on one protein chain and some adjacent sequences modulate further.
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0

A computational AlphaFold screen suggested the KTYQR motif, located N-terminally of the YxF, that appears to also bind STAG1.
27.01.2025 17:34 — 👍 5 🔁 1 💬 1 📌 0
Even though YxF has a large impact on cohesin's LE activity, it doesn't fully account for the impact of the full N-terminal region of CTCF. So we searched more 🔎
27.01.2025 17:34 — 👍 1 🔁 0 💬 1 📌 0

And indeed, FCS experiments showed that STAG1's DNA affinity increases upon addition of the YxF motif.
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0
Curiously, we observed that cohesin also changes direction much less frequently in the presence of YxF! Taking inspiration from Shaltiel et al, Science, 2022, we hypothesized that this is due to a higher DNA affinity of some cohesin subunit in the presence of YxF. Our main candidate: STAG1-kleisin
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0

We started with the known YxF motif. In brief, the YxF motif:
- reduces cohesin's ATPase rate
- reduces LE initiation
- reduces the fraction of complete LE steps (in Magnetic Tweezers)
So these few amino acids alone already tinker quite a lot with cohesin!
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0

Observing LE by Magnetic Tweezers provides a much more detailed view, however. We can see individual steps made by cohesin. Previously, we already observed a plethora of phenomena: cohesin often makes full LE steps, as seen by a height decrease of the magnetic bead. But these are also often reversed
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0

We took the CTCF N-terminal region that is already well-known to interact with cohesin and cut it up into smaller pieces. Then, we exposed cohesin to these various fragments in buffer (no DNA binding domain here!) and observed how cohesin handles that.
27.01.2025 17:34 — 👍 2 🔁 0 💬 1 📌 0
Bridging chemistry, engineering, biology & medicine to advance human health, transform research & train the next generation of scientific leaders at Stanford University.
chemh.stanford.edu
linkedin.com/company/stanford-chem-h
Signup https://bit.ly/44lkPh8
#Climate, #science, #MachineLearning, #SciArt, #Biochemistry, #AI #Media #Art, #Sculpture, #GlassArt
Climate Researcher @Google
Prev: Protein Folding @UW with David Baker, PhD @Bristol
https://fediscience.org/@mtyka
https://www.miketyka.com
Genome Organization & Imaging Researcher | Postdoctoral Fellow at Harvard Medical School | Expert in Advanced Imaging, Image Analysis & Chromatin Biology
https://laurabreimann.github.io/
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
News from the King Lab kinglab.ipd.uw.edu.
Part of @uwmedicine.bsky.social and @uwproteindesign.bsky.social.
Some assembly required.
Our vision is a world where interdisciplinary science flourishes without limit, driving innovation and accelerating discoveries to benefit the world. schmidtsciencefellows.org
Bold science, deep and continuous collaborations.
Card carrying Cell Biologist. Prof. at Stanford Biochemistry. Lover of neglected and emerging model systems. straightlab.stanford.edu
Making life beyond cancer a reality. fredhutch.org
Open source, open science, AI in science for earth/ice and healthcare. IPython creator, @projectjupyter.bsky.social and 2i2c.org co-founder.
Prof @ UC Berkeley Stats, director of @ucbids.bsky.social, co-director @schmidtdse.bsky.social; LBL scientist.
Postdoc at #CDlab, TU Delft | Future PI at Utrecht University | Former PhD at #Dietzlab | DNA origami | Life at the nanoscale | Science & science-fiction
Computational & Genomics Lab studying 3D genome function and dynamics @ Netherlands Cancer Institute
Chromosome biology and genome stability | lab started 2023 @maxperutzlabs.bsky.social @vienabiocenter, Vienna, Austria
Investigating host-pathogen interactions across all domains of life 🌱🦠. Current focus is plants within @pcronald.bsky.social lab at @ucdavis.bsky.social. PhD in Joe Bondy-Denomy’s lab at UCSF. Website: https://erin.phd. Schmidt Science Fellow.
Building a Synthetic Cell (BaSyC) is a research programme with the goal of building an autonomous, self-reproducing synthetic cell.
www.basyc.nl
Single Molecule Biophysics of spatiotemporal chromatin, proteome, and transcriptome || @xtmolbio on X
We create proteins that solve modern challenges in medicine, technology, and sustainability.
• 2024 Nobel Prize in Chemistry
• University of Washington, Seattle
→ ipd.uw.edu