Thank you @plantphys.bsky.social!
30.09.2025 17:55 — 👍 1 🔁 0 💬 0 📌 0

Library-free data-independent acquisition mass spectrometry enables comprehensive coverage of the cyanobacterial proteome
Single-pot solid phase-enhanced sample preparation (SP3) and trifluoroacetic acid workflows open the door to high-throughput quantitative proteomics in cya
Very proud to share the latest development on our (w/ @synbiojazz.bsky.social) mission to bring fast and accessible methods to cyanobacterial proteomics.
Major highlight: 85% Synechococcus proteome coverage in a single shotgun experiment.
Out now in Plant Physiology!
doi.org/10.1093/plph...
19.09.2025 08:20 — 👍 5 🔁 2 💬 0 📌 1
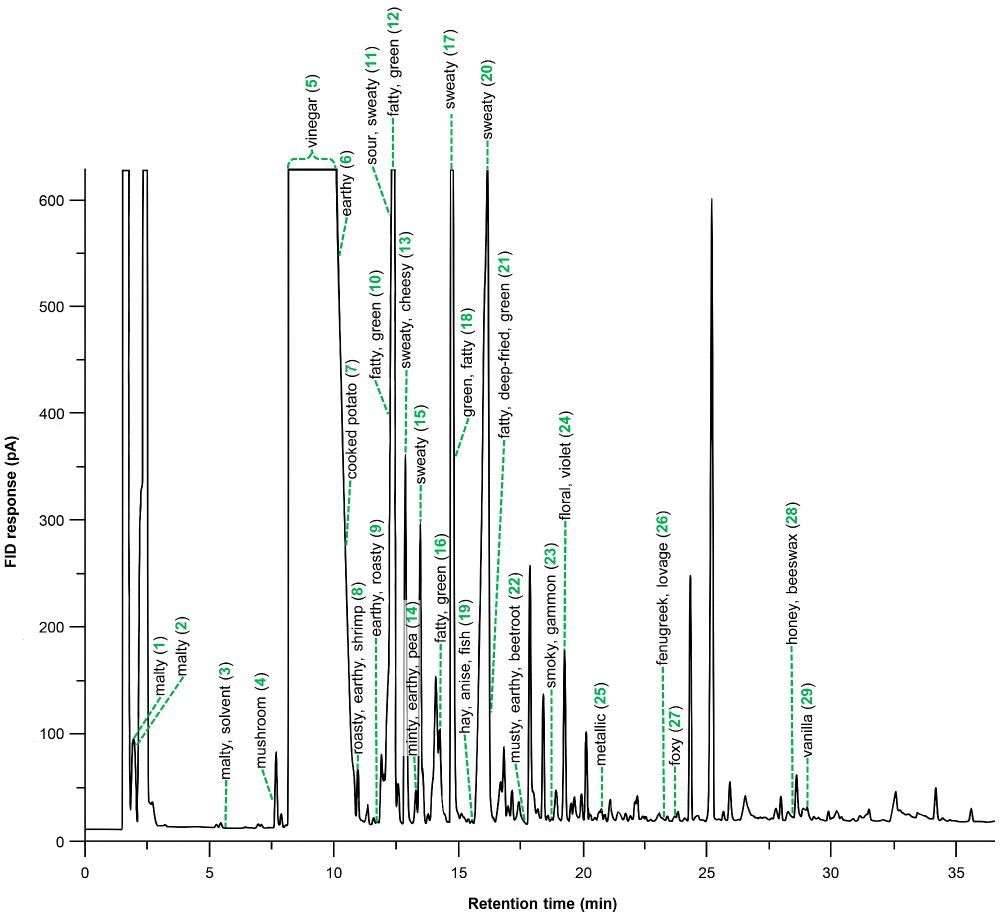
Our new paper on the aroma of #Spirulina is out now in IJMS! A #sensomics approach to find out what makes it smell so unique. Congrats to Katerina Paraskevopoulou, M. Steinhaus, and V. Mall! A great collab between LSB at TUM and NCSR Demokritos. @mdpiopenaccess.bsky.social
doi.org/10.3390/ijms...
16.07.2025 05:30 — 👍 7 🔁 4 💬 0 📌 0
#metabolomics #CompMetabolomics #Visualization #data #organization #annotation #prioritization
26.06.2025 11:48 — 👍 8 🔁 2 💬 0 📌 0
RNA-binding proteins and photosynthesis: The RRM domain–containing protein Rbp3 interacts with ribosomes and the 3’ ends of mRNAs encoding photosynthesis proteins | PNAS www.pnas.org/doi/10.1073/...
26.06.2025 13:59 — 👍 6 🔁 3 💬 2 📌 0
I'm excited to start my independent research group on Bioelectricity in Plant Sciences at the University of Turku, Finland, with an Emerging Investigator grant from the Novo Nordisk Foundation. I will open funded positions in Autumn 2025 to join from Spring 2026. Please email if you see a match.
03.06.2025 11:53 — 👍 9 🔁 2 💬 2 📌 0
That's a fancy video. I was certainly surprised to see the Very Important Protein in Plants discovered in a tropical jungle🌴😅. It's pure marketing ofc (The Krios: Now in Black), but I do like the messaging about discovering the molecular world all around us-- including plants and cyanobacteria🧪🧶🧬🔬🌾🌊
03.04.2025 11:45 — 👍 23 🔁 2 💬 4 📌 1

Substrate-induced assembly and functional mechanism of the bacterial membrane protein insertase SecYEG-YidC
The universally conserved Sec translocon and the YidC/Oxa1-type insertases mediate biogenesis of alpha-helical membrane proteins, but the molecular basis of their cooperation has remained disputed over decades. A recent discovery of a multi-subunit insertase in eukaryotes has raised the question about the architecture of the putative bacterial ortholog SecYEG-YidC and its functional mechanism. Here, we combine cryogenic electron microscopy with cell-free protein synthesis in nanodiscs to visualize biogenesis of the polytopic membrane protein NuoK, the subunit K of NADH-quinone oxidoreductase, that requires both SecYEG and YidC for insertion. We demonstrate that YidC is recruited to the back of the translocon at the late stage of the substrate insertion, in resemblance to the eukaryotic system, and in vivo experiments indicate that the complex assembly is vital for the cells. The nascent chain does not utilize the lateral gate of SecYEG, but enters the lipid membrane at the SecYE-YidC interface, with YidC being the primary insertase. SecYEG-YidC complex promotes folding of the nascent helices at the interface prior their insertion, so the examined cellular pathway follows the fundamental thermodynamic principles of membrane protein folding. Our data provide the first detailed insight on the elusive insertase machinery in the physiologically relevant environment, highlight the importance of the nascent chain for its assembly, and prove the evolutionary conservation of the gate-independent insertion route. ### Competing Interest Statement The authors have declared no competing interest. Deutsche Forschungsgemeinschaft, https://ror.org/018mejw64, Ke1879/3, 267205415 (CRC 1208) European Research Council, https://ror.org/0472cxd90, CRYOTRANSLATION
Very special feelings to announce this one... A project that started like 10 years ago is reaching the finish line, ready to shine. In a dream-team with @beckmannlab.bsky.social we solved the long-chased structure of the active membrane protein insertase SecYEG-YidC
www.biorxiv.org/content/10.1...
27.05.2025 09:21 — 👍 59 🔁 24 💬 7 📌 7

High-performance proteomics at any chromatographic flow rate
Current applications of mass spectrometry-based proteomics range from single cell to body fluid analysis that come with very different demands regarding sensitivity or sample throughput. Additionally, the vast molecular complexity of proteomes and the massive dynamic range of protein concentrations in these biological systems require very high-performance chromatographic separations in tandem with the high speed and sensitivity afforded by mass spectrometer. In this study, we focussed on the chromatographic angle and, more specifically, systematically evaluated proteome analysis performance across a wide range of chromatographic flow rates (0.3 – 50 μL/min) and associated column diameters using a Vanquish Neo UHPLC coupled online to a Q Exactive HF-X mass spectrometer. Serial dilutions of HeLa cell line digests were used for benchmarking and total analysis time from injection-to-injection was intentionally fixed at 60 minutes (24 samples per day). The three key messages of the study are that i) all chromatographic flow rates are suitable for high-quality proteome analysis, ii) capLC (1.5 μL/min) is a very robust, sensitive and quantitative alternative to nanoLC for many applications and iii) showcased proteome, phosphoproteome and drug proteome data provide sound empirical guidance for laboratories in selecting appropriate chromatographic flow rates and column diameters for their specific application.
### Competing Interest Statement
R.Z. and C.P. are employees of Thermo Fisher Scientific. BK is a non-operational co-founder and shareholder of MSAID. The other authors declare no competing interests.
* ABBREVIATIONS
:
ACN
: Acetonitrile
CAA
: Chloroacetamide
capLC
: Capillary Liquid Chromatography
CSF
: Cerebrospinal Fluid
CV
: Coefficient of Variation
DDA
: Data-dependent Acquisition
DIA
: Data-Independent Acquisition
DMSO
: Dimethyl Sulfoxide
DTT
: Dithiothreitol
EC50
: Effective Concentration to Reduce 50% of Protein Binding to Kinobeads
FBS
: Fetal Bovine Serum
fmol
: Femtomole
FDR
: False Discovery Rate
FWHM
: Full Width at Half Maximum
HeLa
: Human Cervical Carcinoma Cell Line
HLB
: Hydrophilic-Lipophilic Balance
HPLC
: High-Performance Liquid Chromatography
i.d.
: Internal Diameter
Kdapp
: Apparent Dissociation Constant
LC
: Liquid Chromatography
LC-MS/MS
: Liquid Chromatography-Tandem Mass Spectrometry
µg
: Microgram
min
: Minute
µL
: Microliter
µLC
: Microflow Liquid Chromatography
mm
: Milimeter
mM
: Milimolar
MS
: Mass Spectrometry
ng
: Nanogram
nL
: Nanoliter
nLC
: Nanoflow Liquid Chromatography
nM
: Nanomolar
PSM
: Peptide-spectrum Match
RT
: Retention Time
SPD
: Samples per Day
SWATH-MS
: Sequential Window Acquisition of All Theoretical Mass Spectra
TFA
: Trifluoroacetic Acid
Å
: Angstrom
Which flow rate is most suitable for MS-based #proteomics and #chemoproteomics?
We have been thinking about this for a long time (nanoLC, capLC, µLC). According to a new @biorxivpreprint.bsky.social by the @kusterlab.bsky.social, it seems like all options work very well. www.biorxiv.org/cont...
22.04.2025 06:00 — 👍 13 🔁 2 💬 1 📌 1
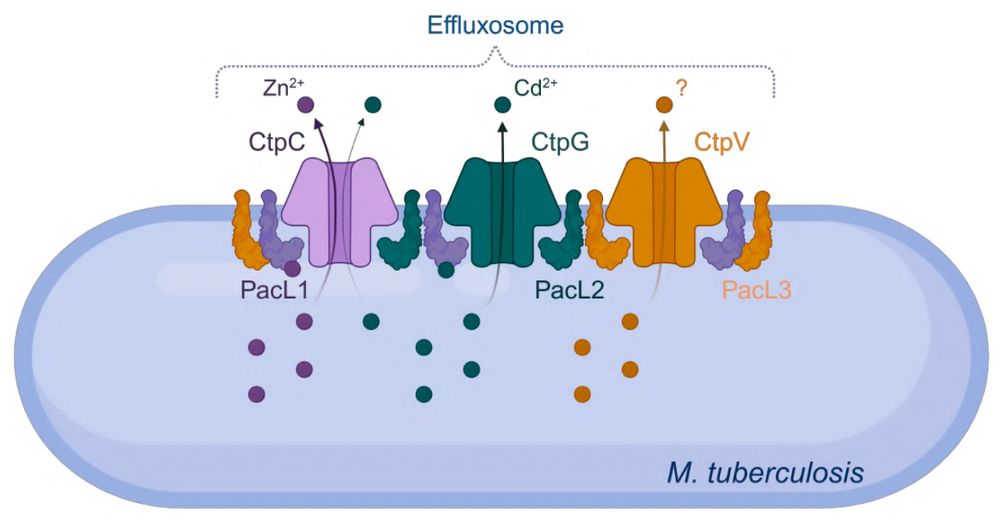
Shout-out to @pierredupuy.bsky.social and colleagues!
Excited to share our latest study, where we uncovered a previously unknown bacterial defense system in M. #tuberculosis: effluxosomes — dynamic membrane clusters that coordinate resistance to multiple toxic metals.
shorturl.at/Ek0HD
#MicroSky
27.03.2025 06:09 — 👍 62 🔁 32 💬 9 📌 2
A story of cyanobacterial symbiosis with a twist! Take a look at our recent paper to learn more about the unexpected relationship of Nostoc and Agrobacterium #cyanobacteria #symbiosis #proteomics
11.03.2025 10:40 — 👍 0 🔁 0 💬 0 📌 0
Proud to be a part of this work. If you want to improve the expression and activity of your favourite P450 check this out!
06.03.2025 16:11 — 👍 3 🔁 0 💬 0 📌 0
Me and @synbiojazz.bsky.social please
06.03.2025 15:29 — 👍 1 🔁 0 💬 1 📌 0
DeepSeMS: a large language model reveals hidden biosynthetic potential of the global ocean microbiome www.biorxiv.org/content/10.1... #jcampubs
04.03.2025 17:00 — 👍 6 🔁 4 💬 0 📌 0
Head of Molecular Microbiology Dept @JohnInnesCentre.bsky.social. Natural products & Streptomyces interactions with plants and insects. Drummer, dad, Norwich City
http://streptomyces.org.uk for protocols & resources. Engagement with @sawtrust.bsky.social
Director of Protea Glycosciences
Accelerating glycoscience research
Doctoral Program in Biology University of Tsukuba, interested in fatty acids metabolism of cyanobacteria/光合成若手の会幹事/トビタテ14期理系コース🇫🇮/学芸員資格/野生動物研究会元広報担当・元ARE生
https://plmet.biol.tsukuba.ac.jp/
Pastel BioScience | t.ly/y4tAy
Proteomics conferences here >>> t.ly/GSBz-
Proteomics databases and scripts here >>> t.ly/iNYXg
Microbiologist exploring plant microbial interactions | Rhizosphere engineering |Symbiosis #Mycorrhiza #Rhizobium #legumes #postdoctoral_research_associate 🇺🇸
Researcher from 🇫🇷 doing #cryoEM ❄️🔬 at Uppsala University 🇸🇪
Working on cyanobacterial carboxysomes 🦠, still interested in nucleosome 🧬🙃🧬
Mostly […]
[bridged from https://fediscience.org/@Guillawme on the fediverse by https://fed.brid.gy/ ]
Assistant Professor in Plant Biotech at Lancaster Environment Centre, UK.
Interested in Rubisco, photosynthesis, plant factories, plant synbio.
Consumer of Tim Tams and AU/PT wine.
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
Assistant Professor of Biology at Boston University ::: Science Journalist / Communicator ::: National Geographic Explorer
Group Leader @ The John Innes Centre.
Photosynthetic Gene Expression 🌿🧬 |
Cryo-EM
We investigate the genetic regulation and physiology of cyanobacteria. Our research is focused in the identification of regulatory networks involving FUR proteins and the formation of biofilms.
Researcher, interested in Cyanobacteria and Biotechnology. He/him
Exploring how cyanobacteria changed the wO2rld, One oxygen molecule at a time!
Specifically on the early Earth as well as current day climate change effects on primary productivity (both C & N fixation), symbioses and secondary metabolite production.
Leibniz Institute: Hans Knöll Institute
Jena
The research group Applied Systems Biology is concerned with the mathematical modeling and computer simulation of infection processes caused by human-pathogenic fungi.
University of Edinburgh; Institute of Molecular Plant Sciences; photosynthesis; plant and algal engineering biology; cyanobacterial biotechnology.
Plant scientist. Keen on understanding the inner works of photosynthesis. Works at @hhu.bsky.social
https://www.plant-biochemistry.hhu.de
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
















