⚠️ Upcoming changes:
As of Reactome V94, the following files will no longer be available: “Protégé 2.0 ontology files”, and “Events in the BioPAX level 2 format”. If you require these files for your work, please let our team know via help@reactome.org.
24.06.2025 16:48 — 👍 0 🔁 0 💬 0 📌 0
⚠️ Reminder:
As of Reactome V93, our Docker images are no longer available on DockerHub and have been migrated to AWS Elastic Container Registry (ECR). Users should pull images from AWS ECR moving forward.
24.06.2025 16:48 — 👍 0 🔁 0 💬 1 📌 0
Community - Reactome Pathway Database
Reactome is pathway database which provides intuitive bioinformatics tools for the visualisation, interpretation and analysis of pathway knowledge.
👏 Thank you to our contributors, authors who have provided expert text and guidance, and peer-reviewers who provide feedback and ensure that everything entering Reactome is accurate. If you are interested in contributing, learn more about how to do so at this link: reactome.org/community
24.06.2025 16:48 — 👍 0 🔁 0 💬 1 📌 0
Reactome Strengthens Accuracy by Monitoring for Retracted Publications - Reactome Pathway Database
Reactome is pathway database which provides intuitive bioinformatics tools for the visualisation, interpretation and analysis of pathway knowledge.
Reactome audits all curated refs against Retraction Watch. Of 40,000+ papers, just 70 were retracted — and all annotations reviewed or removed. A small number, but we take every one seriously. #FAIRdata #OpenScience #Reactome
🖥️🧬
Read more about it here: reactome.org/content/reac...
30.05.2025 14:18 — 👍 5 🔁 2 💬 0 📌 0
Reactome Recognized with CoreTrustSeal Certification - Reactome Pathway Database
Reactome is pathway database which provides intuitive bioinformatics tools for the visualisation, interpretation and analysis of pathway knowledge.
Reactome is now CoreTrustSeal certified, meaning that our pathway data meets global standards for transparency, accessibility & long-term stability. Learn more at reactome.org/about/news/2...
A big win for #FAIRdata, open science, and everyone who relies on Reactome for biological knowledge.
🖥️🧬
22.05.2025 13:41 — 👍 10 🔁 5 💬 0 📌 0
Check out this recap, where our #Reactome Outreach Coordinator summarizes some of the team discussions we had at #ReactomeSAB 2025!
🖥️🧬
20.05.2025 15:04 — 👍 2 🔁 0 💬 0 📌 0
No matter where you are in your career— grad student, postdoc, or early-career researcher—you can play a meaningful role in advancing open science and shaping the Reactome knowledgebase.
07.04.2025 17:38 — 👍 0 🔁 0 💬 0 📌 0
Reactome is committed to FAIR data principles, meaning your contributions don’t just stay in one place—they’re disseminated globally and reused in a variety of biomedical contexts, from drug discovery to systems biology.
07.04.2025 17:38 — 👍 1 🔁 0 💬 1 📌 0
In recognition of their work, contributors receive:
✅ ORCID credit linked to their profile
✅ A citable DOI for each pathway, perfect for CVs and grant applications
✅ Impact—your work becomes part of a resource used by thousands of researchers and integrated into leading bioinformatics platforms.
07.04.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
More than 900 expert scientists have contributed to Reactome by authoring or peer-reviewing pathways, helping ensure the quality and accuracy of this freely available knowledgebase.
07.04.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
YouTube video by Reactome Knowledgebase
Contributing to Reactome
Check out our new Youtube video about contributing to Reactome!
Learn more by watching our youtube video, created by Cristoffer Sevilla!
www.youtube.com/watch?v=P5Gi...
Like and subscribe to our Youtube channel for more updates!
🖥️🧬
07.04.2025 17:38 — 👍 3 🔁 3 💬 1 📌 0
⚠️ In our next version (v93) of Reactome, we will be migrating our Docker image hosting from DockerHub to AWS Elastic Container Registry (ECR). Users will be able to pull our container images from AWS ECR. Please reach out to help@reactome.org if you have any questions.
🖥️🧬
02.04.2025 15:19 — 👍 0 🔁 0 💬 0 📌 0
A sincere thank you to our external contributors, who provide valuable feedback and review for our pathways. If you are interested in contributing, check out how you can do so, at this link: reactome.org/community
🖥️🧬
02.04.2025 15:19 — 👍 0 🔁 0 💬 1 📌 0
Reactome Release News: V92 of #Reactome is live! 🎉
We have exciting new and updated topics and pathways in this release. This release, we added 81 new reactions to Reactome, which you can read about at this link: reactome.org/about/news/2...
🖥️🧬
02.04.2025 15:19 — 👍 1 🔁 1 💬 1 📌 0
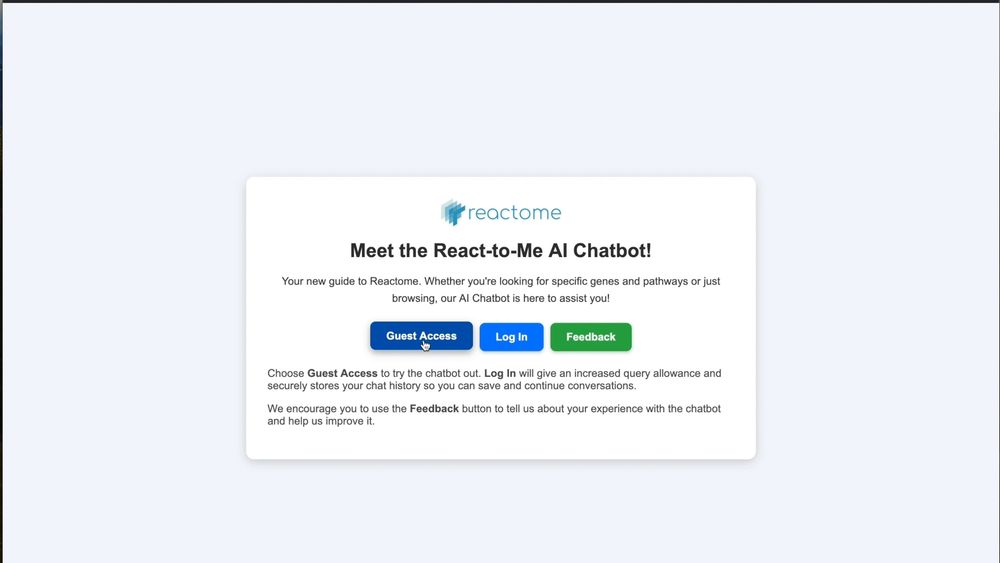
GIF showing interacting with the Reactome AI chatbot. The chatbot gives a quick explanation of ECM signaling and offers clickable links to Reactome's knowledgebase.
Discover Pathways with Ease!
Explore biological pathways like never before with the React-to-Me AI Chatbot. We’d love your feedback to improve it! Try it now and share your experience through our short questionnaire. Link: reactome.org/chat/
🧬🖥️ 🧪
14.02.2025 18:01 — 👍 7 🔁 6 💬 0 📌 0

GIF showing interacting with the Reactome AI chatbot. The chatbot gives a quick explanation of ECM signaling and offers clickable links to Reactome's knowledgebase.
Discover Pathways with Ease!
Explore biological pathways like never before with the React-to-Me AI Chatbot. We’d love your feedback to improve it! Try it now and share your experience through our short questionnaire. Link: reactome.org/chat/
🧬🖥️ 🧪
14.02.2025 18:01 — 👍 7 🔁 6 💬 0 📌 0
So glad that you found us! Thanks for posting about our work!
30.01.2025 15:06 — 👍 1 🔁 0 💬 0 📌 0
#ReactomeResearchSpotlight is updated monthly on our website's homepage! Our staff selects candidate articles to feature. After team discussion, the winner is posted on our home page.
Contact us at help@reactome.org with if you want to be featured as the next Spotlight!
13.01.2025 18:59 — 👍 0 🔁 0 💬 0 📌 0
In S2FCD, Oncogene- and Oxidative stress-induced senescence pathways were upregulated and Signaling by TGF-beta receptor complex events were downregulated suggesting distinct subtype-specific therapeutic strategies.
13.01.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
Reactome pathway enrichment analysis revealed subtype-specific dysregulations. In IDICD, the Nuclear receptor transcription factor pathway, Butyrophilin family interactions and Intestinal infectious disease events were upregulated while Cytokine signaling in immune system events were downregulated.
13.01.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
A living organoid biobank of patients with Crohn’s disease reveals molecular subtypes for personalized therapeutics - Reactome Pathway Database
Reactome is pathway database which provides intuitive bioinformatics tools for the visualisation, interpretation and analysis of pathway knowledge.
The #ReactomeResearchSpotlight for January is a paper by Tindle et al. that identified two Crohn’s disease (CD) molecular subtypes - immune-deficient infectious CD and stress and senescence-induced fibrostenotic CD - through multi-omics and functional analyses of patient-derived organoids
🖥️🧬
13.01.2025 18:59 — 👍 0 🔁 0 💬 1 📌 0
🔓Juan Antonio Vizcaino on open data practices in proteomics, highlighting data reuse activities in the context of big data approaches
💻Eliot Ragueneau demonstrating @reactome.org GSA for multi-omics data analysis
#FoG2025
09.01.2025 12:17 — 👍 5 🔁 2 💬 1 📌 0
Release News: V91 of #Reactome is live! 🥂 🎉
We have exciting new and updated topics and pathways in this release, which you can read about at this link: reactome.org/about/news/2...
19.12.2024 17:56 — 👍 1 🔁 4 💬 1 📌 0
Please consider spending a few moments of your day supporting our resource by providing your feedback to our team! The things we learn from the user survey is essential for our continued success!
🖥️🧬
15.11.2024 16:13 — 👍 1 🔁 4 💬 0 📌 0

Let's play 20 questions!
If Reactome has touched your work or helped you save valuable time, please consider giving us 5 minutes of your time through our user survey, reachable at our home page, or at the following link: docs.google.com/forms/d/e/1F...
🧪🖥️🧬
01.11.2024 18:12 — 👍 3 🔁 4 💬 0 📌 1
YouTube video by Reactome Knowledgebase
Reactomes Downloadable materials
As an #opensource and #public resource, we offer numerous ways for our community to re-use any and every part of the content we create. All you need to do is cite us for our work, so others can also easily find these useful resources.
🖥️🧬
26.11.2024 18:02 — 👍 0 🔁 0 💬 0 📌 0
dockstore.org is a free and open source platform for sharing reusable and scalable analytical tools and workflows
The Ensembl project seeks to enable genomic science by providing high-quality, integrated annotation.
Vertebrates: www.ensembl.org
Non-vertebrates: www.ensemblgenomes.org
You can test the new Ensembl browser and share your feedback at beta.ensembl.org
⚛️ Postdoc in Computational Biomedicine @saezlab.bsky.social @Heidelberg
🧪 Facinated by Cancer Metabolism, Gene Regulation & Multiomics
🎓 Previously @frezzalab.bsky.social @Cambridge_Uni and @Wuerzburg_Uni
🧫 Love to paint Scienceart
EMBL's European Bioinformatics Institute (EMBL-EBI) provides open biological data resources and tools, and performs basic research in computational biology. https://www.ebi.ac.uk/
I’m a computational biologist and biochemist working on synthetic tumor genome generation 💻🧬 Postdoctoral researcher at University of Toronto (UofT) and Ontario Institute for Cancer Research (OICR)
Not-for-profit publisher of Development, Journal of Cell Science, Journal of Experimental Biology, Disease Models & Mechanisms and Biology Open. Host of community sites the Node, preLights and FocalPlane. Supporting biologists and inspiring biology.
This is the official account for the Society for Developmental Biology. Skeets on all the latest Dev Bio news, meetings and science! www.sdbonline.org
Bridging Research and Education with Model ORganisms. We are an international grassroots organization of scientists with the mission to build a network dedicated to increasing experiential learning for undergraduate biology students. brewmor.org
Est. 1929, JAX is a non-profit scientific research institute specializing in genetics, genomics & mouse models of disease.
USA, Japan, China: https://www.jax.org/
Advancing research and teaching in the most dynamic and vital areas of biological research
The Gene Ontology (GO, geneontology.org) knowledgebase is the world’s largest source of information on gene function. Our mission is to develop a comprehensive, computational model of biological systems, ranging from the molecular to organism level.
Curating and cataloging the world's data for the fruit fly Drosophila melanogaster into our database at http://flybase.org. Aiding discovery since 1992.
The Alliance of Genome Resources supports biological science via data integration, standardization of data models and interfaces, and unified outreach.
Find us at https://www.alliancegenome.org.
Cognitive/social scientist and occasional coder. Purveyor of Jurassic Park memes. Writes on modernization, technology, and economic democracy. Consciously uncoupling from corporate shenanigans. Halifax, Nova Scotia
https://morungos.com/
Head of Protein Sequence Resources at EMBL-EBI.
Professor passionate about student success & team science #firstgen #Brassica enthusiast walking Dogs of the Plant World
#Agriculture #Food #HigherEd #EduSky #Agsky #SciComm & more
#Soil #Crop #Science ; opinions mine
Asst Prof of Pharmaceutical Sciences #UCIrvine | Studying #microprotein and #peptide biology in cancer and metabolism | #FirstGen | #LAnative | 🇵🇸🇲🇽-🇺🇸 | formerly @tfmphd on the birdapp | https://faculty.sites.uci.edu/martinezlab/
Developing macromolecular structure resources and tools for life science.
- Founding partner of the wwPDB.
- Manage the community-led PDBe-KB resource and the AlphaFold Protein Structure Database, a collaboration with Google DeepMind.
- Part of EMBL-EBI
Marketing and Comms Manager for EMBL-EBI Training. I'll be here to tell you about all our wonderful #bioinformatics training opportunities. All messages are my own views and not affiliated or endorsed by EMBL and EMBL-EBI.
www.ebi.ac.uk/training