Super excited about first Shendure/Baker Lab collaboration & preprint on a multiplex sequencing-based strategy for screening de novo proteome editors in mammalian cells. Kudos to the brilliant Chase Suiter (not here) & @greenahn.bsky.social on the work! Preprint here:
www.biorxiv.org/content/10.1...
14.10.2025 18:44 — 👍 97 🔁 33 💬 0 📌 0
We're hiring to expand on the work to understand the human genome by engineering it!
lnkd.in/da-gitNc
10.02.2025 08:53 — 👍 12 🔁 8 💬 0 📌 0
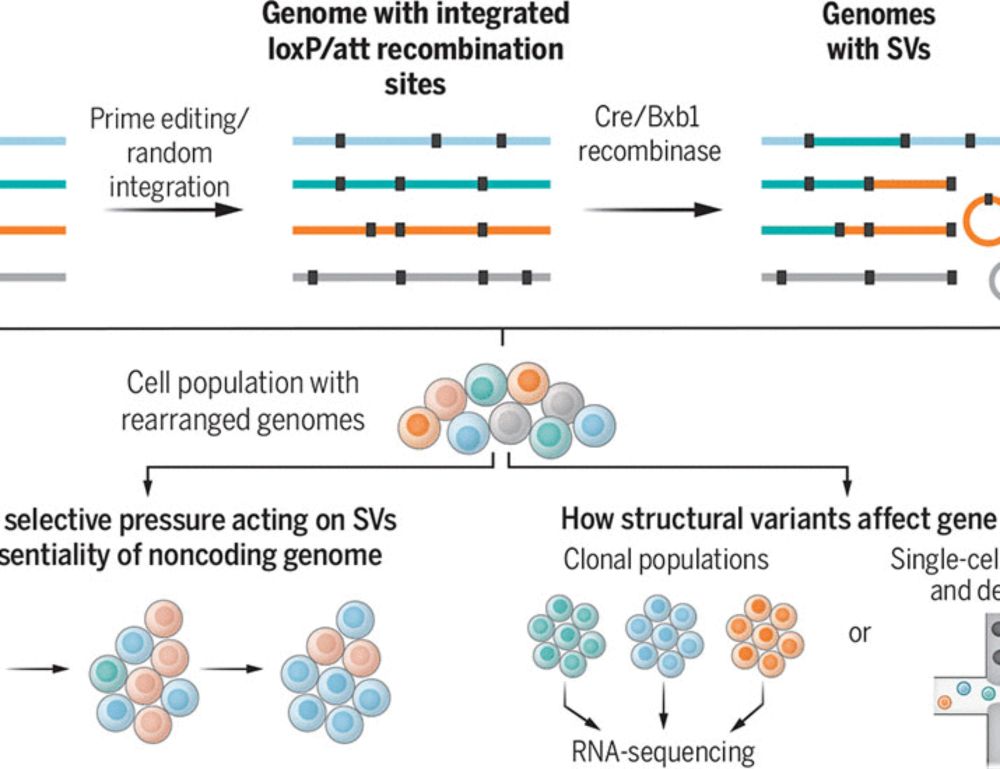
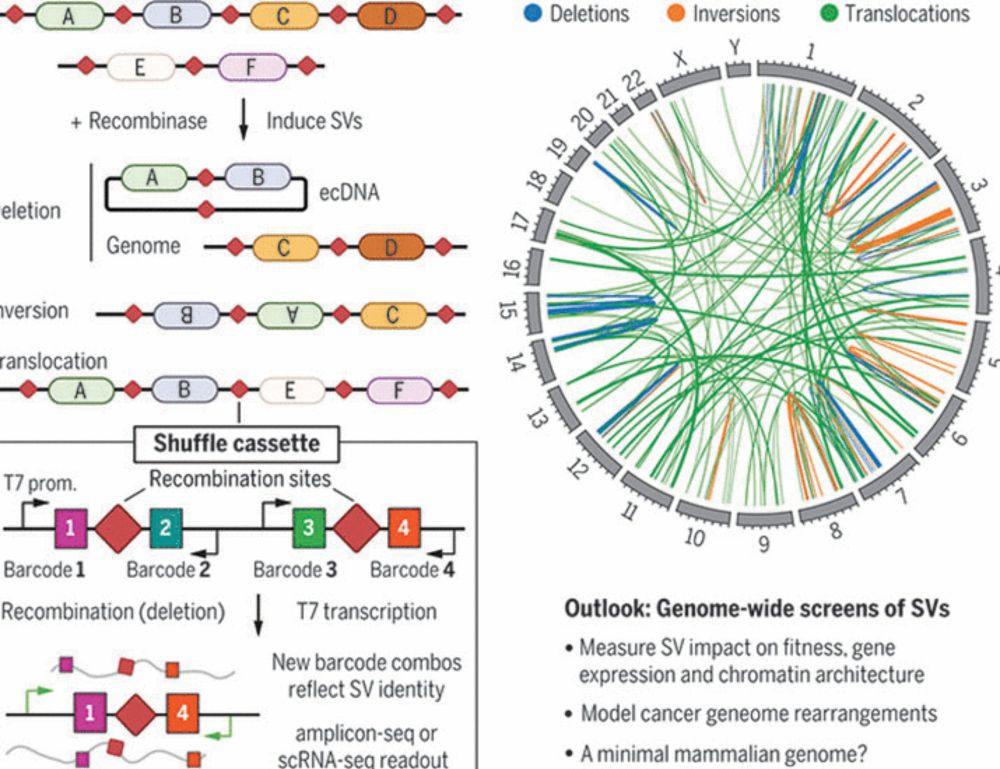
Genome recombination on demand
Large genome rearrangements in mammalian cells can be generated at scale
@science.org Genome recombination on demand | Science www.science.org/doi/10.1126/... a Perspective by @seczmarta.bsky.social Lars Steinmetz @stanford.edu on two studies bit.ly/4hzFRMg + bit.ly/4jyT4Hf that generate large genome rearrangements in mammalian cells @ unprecedented scale #synbio #genome
01.02.2025 18:58 — 👍 48 🔁 20 💬 1 📌 0
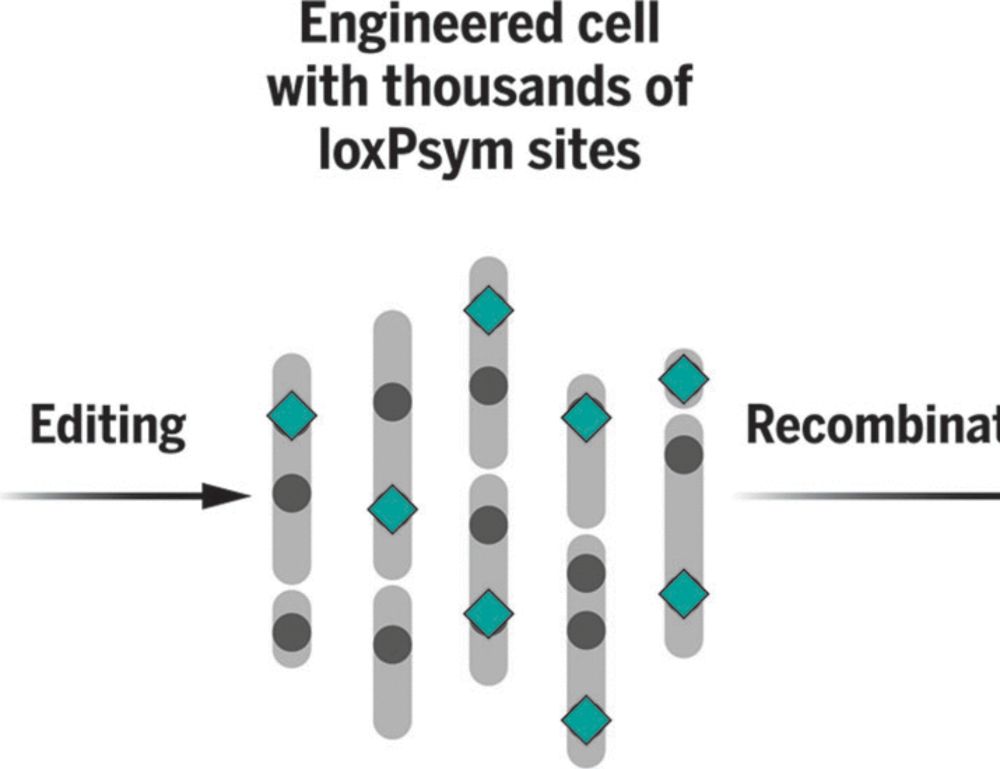
🧬@science.org Randomizing the human genome by engineering recombination between repeat elements bit.ly/4jyT4Hf @jonaskoeppel.bsky.social @f-raphael.bsky.social @geochurch.bsky.social @proftomellis.bsky.social @leopoldparts.bsky.social +al. @sangerinstitute.bsky.social @harvardmed.bsky.social #synbio
01.02.2025 18:48 — 👍 10 🔁 5 💬 0 📌 0
https://www.pinglay-lab.com/
If tinkering with genomes - designing, writing, shuffling and augmenting them excites you, come join us! We are hiring at all levels.
www.pinglay-lab.com
31.01.2025 19:41 — 👍 1 🔁 1 💬 1 📌 0
Nothing much to add here from my side! It was in fact a very similar situation with @f-raphael.bsky.social and us. We connected for a different reason and figured out we both tried to insert loxP sites into LINE1 and then decided to join forces
31.01.2025 20:37 — 👍 4 🔁 0 💬 0 📌 0
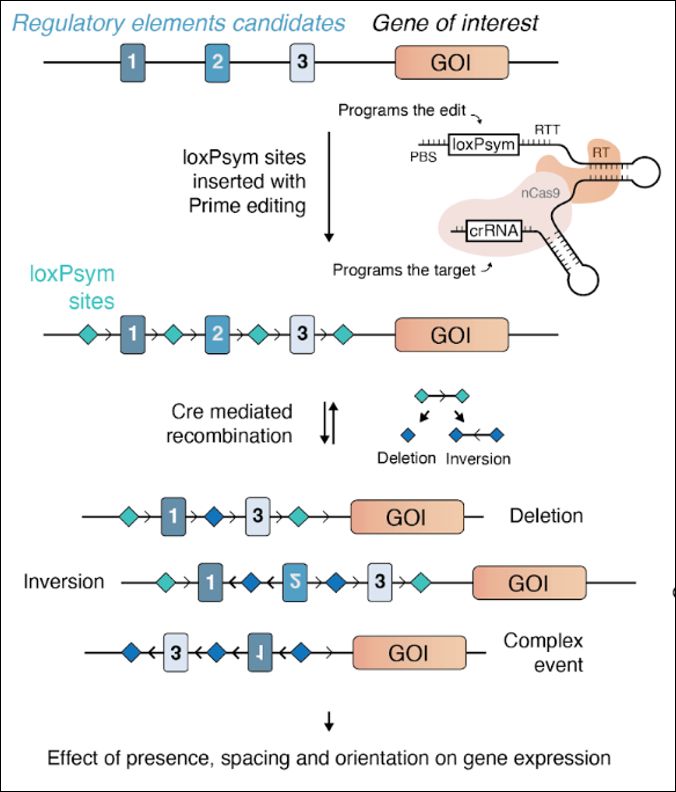
Enhancer scrambling strategy
We are happy to share our enhancer scramble story, a strategy to create hundreds of stochastic deletions, inversions, and duplications within mammalian gene regulatory regions and associate these new architectures with gene expression levels 🧵
www.biorxiv.org/content/10.1...
15.01.2025 20:32 — 👍 183 🔁 77 💬 3 📌 2
Excited to share my latest preprint on establishing a generalizable toolkit for decoding the gene regulatory landscape using two types of CRISPR screens. Big thanks to my amazing mentor @nevillesanjana.bsky.social for the in-depth thread below. Looking forward to feedback and comments!
28.12.2024 20:26 — 👍 10 🔁 2 💬 1 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
Does my mutation have the same impact as yours? Population genetics 🤠 🥸 🤓 🤡 meets single cell CRISPRi ⚡ ! www.biorxiv.org/content/10.1... Led by Claudia Feng, Oliver Stegle, Britta Velten, @sangerinstitute.bsky.social .
02.12.2024 13:57 — 👍 58 🔁 24 💬 2 📌 3
Postdoc @broadinstitute.org @harvard.edu #MGH - Immunogenomics - PhD in Genomics🧬 @cam.ac.uk @sangerinstitute.bsky.social🔬 #LCG-UNAM
We are a research lab at the Netherlands Cancer Institute. We develop and apply new genomics tools to study genome biology and gene regulation.
Professor at the University of British Columbia @sbmeubc.bsky.social
Specially Appointed Professor at Osaka University PRIMe
https://yachie-lab.org
Exploring #SyntheticBiology & #Bioinformatics + Documenting my learning journey | Built by @noureldenrihan.bsky.social | 🌐 djosergenomics.github.io
Group Leader @ BIH Berlin and MPI Molecular Genetics. I play around with DNA because I‘d like to know how the genome functions.
Virginie Hamel & Paul Guichard Lab at University of Geneva
#cryoEM/ET❄️ and #UExM ⚗️ #ExpansionMicroscopy #TeamTomo
Genève, Suisse 🇨🇭
https://mocel.unige.ch/research-groups/guichard-hamel/overv
Grad student advised by Jay Shendure and Nobu Hamazaki. NSF GRFP fellow
MD/PhD Univ Wash, PhD candidate in Jay Shendure lab | Yale '18 | NRSA Fellow | genomics & precision medicine | she/her | views & opinions are my own 🧬 💫
Nobuhiko (Nobu) Hamazaki, Ph.D. Assistant prof. @University of Washington (UW). Stem cell engineering, germ cell and embryonic development.
BioF:GREAT is an NSF BioFoundry located at UGA's Complex Carbohydrate Research Center focused on democratizing glycoscience by increasing accessibility to resources, education, and training.
Mexican Historian & Philosopher of Biology • Postdoctoral Fellow at @theramseylab.bsky.social (@clpskuleuven.bsky.social) • Book Reviews Editor for @jgps.bsky.social • https://www.alejandrofabregastejeda.com • #PhilSci #HistSTM #philsky • Escribo y edito
Scientific content aggregator with custom content feeds. Science news, jobs, events and more. Sign up at https://www.scientific.today.
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Scientist and medical doctor. Biology AI/ML methods, gene regulation, DNA sequence models, single cells. Doing a PhD in computational biology at @molgen.mpg.de.
Associate professor @ South China University of Technology. Interested in synthetic biology, genome editing, and directed evolution.
scientist | pacifist | transposable elements | evo-devo
Former PhD @trono-lab.bsky.social @ EPFL
Postdoc @ Jachowicz lab @imbavienna.bsky.social
Science driven evidence, biotechnology, synthetic biology, health, cancer, alternative food
The opinions expressed are those of the individual alone and do not reflect those of EISMEA, the EIC or the European Commission.