If you like to sample from the Boltzmann distribution and are in San Diego for NeurIPS, be sure to check out Rishal's (@rishalchich.bsky.social) poster (#2110). Great work with Nick Boffi (@nmboffi.bsky.social) and Jacky Chen.
neurips.cc/virtual/2025...
arxiv.org/abs/2507.00846
05.12.2025 18:26 — 👍 11 🔁 2 💬 0 📌 0
Our new preprint PharmacoForge: Pharmacophore Generation with Diffusion Models is out now! PharmacoForge quickly generates pharmacophores for a given protein pocket that identify key binding features and find useful compounds in a pharmacophore search. Check it out! 🧪 doi.org/10.26434/che...
27.05.2025 19:11 — 👍 21 🔁 9 💬 1 📌 0

Happy to introduce 🔥LaM-SLidE🔥!
We show how trajectories of spatial dynamical systems can be modeled in latent space by
--> leveraging IDENTIFIERS.
📚Paper: arxiv.org/abs/2502.12128
💻Code: github.com/ml-jku/LaM-S...
📝Blog: ml-jku.github.io/LaM-SLidE/
1/n
22.05.2025 12:24 — 👍 7 🔁 8 💬 1 📌 1

Long & windy road of academic publishing! Few journal rejections and two years (!!!) after preprint, AIMNet2 paper was just published @chemsocrev.rsc.org With 69 citations to it as of now, it's immediately part of 2025 HOT🌶️ Article collection. pubs.rsc.org/en/content/a... #chemsky #compchem
29.04.2025 17:54 — 👍 52 🔁 9 💬 3 📌 1

Thanks to everyone for helping make #ICLR2025 successful. 🥳 It was an honor to serve as General Chair.
The best part was using @iclr-conf.bsky.social as my personal meme account the past two years (first as Senior Program Chair). I hope that future ICLR organizers continue the tradition. 🤪
28.04.2025 10:31 — 👍 29 🔁 3 💬 2 📌 0
Please welcome AITHYRA, the Research Institute for Artificial Intelligence of the Austrian Academy of Science on social media. Follow us and connect via Bluesky and LinkedIn 👋
28.04.2025 07:45 — 👍 17 🔁 5 💬 0 📌 0

Metagenomic-scale analysis of the predicted protein structure universe
Protein structure prediction breakthroughs, notably AlphaFold2 and ESMfold, have led to an unprecedented influx of computationally derived structures. The AlphaFold Protein Structure Database now prov...
Our latest preprint is out on bioRxiv!
A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.
28.04.2025 11:16 — 👍 39 🔁 13 💬 2 📌 0
Hello, we are AITHYRA!
I am very excited to share with you the corporate identity of AITHYRA, the Research Institute for Biomedical AI.
Check out the brand design video (sound on!) to learn more.
**REMINDER**
One week left to apply for Starting PI positions (Life Science and AI/ML). Come join us!
28.04.2025 08:38 — 👍 20 🔁 8 💬 0 📌 0
"De novo prediction of protein structural dynamics"
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
27.04.2025 14:16 — 👍 70 🔁 17 💬 5 📌 1

Sara with .5 mode selfie (thanks kids) in front of aacr sign
Who is at #AACR25? I’m here representing my group’s work at the intersection of AI and drug resistance in cancer, and want to hear about what you’re doing!(1/4)
28.04.2025 13:26 — 👍 8 🔁 1 💬 3 📌 0
@iclr-conf.bsky.social blog posts are now live at iclr-blogposts.github.io/2025/blog! Unfortunately I won't be able to present our blog post at the conference in person 😔, but I am happy to chat about it online if you find it interesting! Poster attached 🙂.
23.04.2025 22:17 — 👍 0 🔁 0 💬 0 📌 0
With this baseline, we emphasize that such generative methods should focus on computational efficiency and transferability in their model design and benchmarks.
Work done with Daniel ( @countrsignal.bsky.social ), Justin ( @shaojus.bsky.social ), and Minhyek ( @minhyekj.bsky.social )
31.03.2025 17:18 — 👍 0 🔁 0 💬 0 📌 0

New "blogpost" from our lab, that got accepted at ICLR 2025! We compare an old MCMC method known as Sequential Monte Carlo to generative models trained on energy functions (iDEM/iEFM) and show that MCMC does better. Check it out here: rishalaggarwal.github.io/ebmvsmcmc/
31.03.2025 16:57 — 👍 4 🔁 4 💬 1 📌 2

AlphaFold3 still tends to overfit regions that AF2 multimer predicts to be disordered. Almost all disordered regions become helices in AF3.
15.12.2024 02:03 — 👍 55 🔁 20 💬 1 📌 2
Going into @workshopmlsb.bsky.social , a few thoughts on the PLINDER dataset. This is a monumental and highly appreciated undertaking for extracting protein-ligand interactions from the PDB. First the good...
15.12.2024 18:52 — 👍 55 🔁 15 💬 4 📌 2
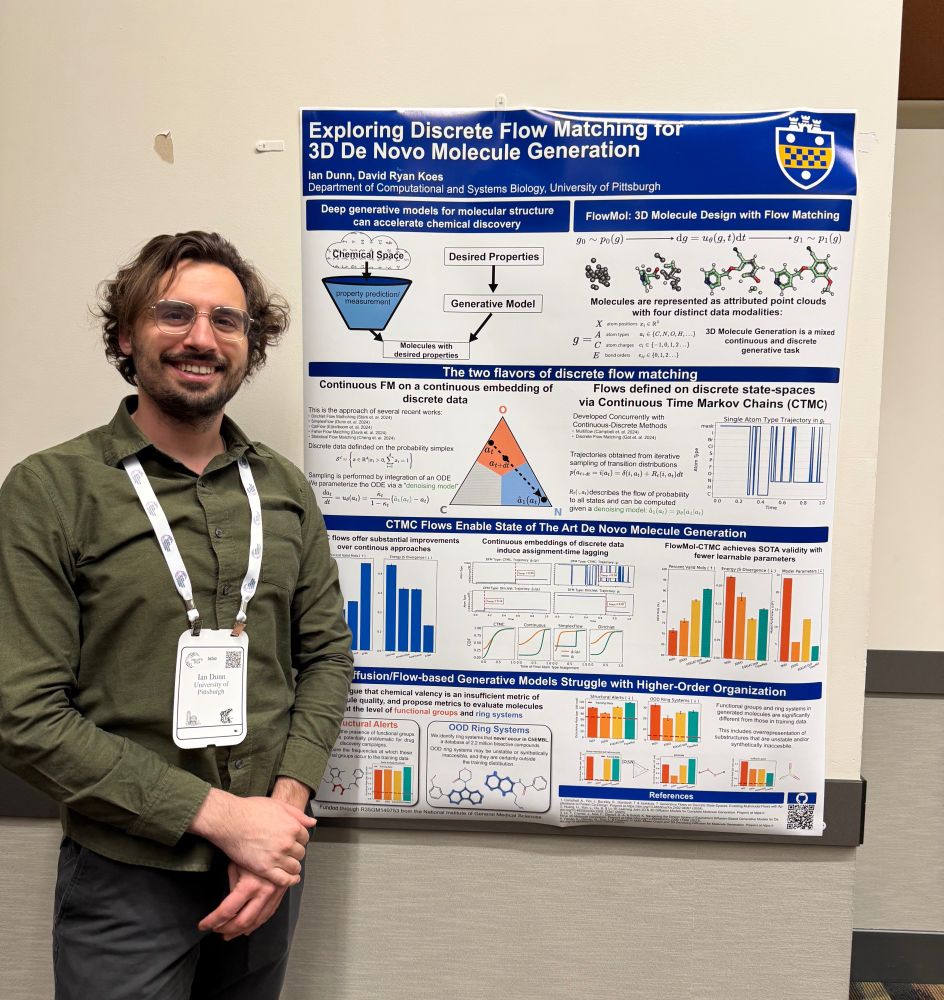
I'm presenting a new paper "Exploring Discrete Flow Matching for 3D De Novo Molecule Generation" at @workshopmlsb.bsky.social this week! More info in this thread but reach out if want to chat at NeurIPS about generative models or molecular design. arxiv.org/abs/2411.16644
11.12.2024 21:21 — 👍 51 🔁 12 💬 2 📌 3
More excellent work from the indomitable @ian-dunn.bsky.social Come check it out at @workshopmlsb.bsky.social
11.12.2024 22:20 — 👍 7 🔁 1 💬 0 📌 0
Excited about BioEmu?
Opening this position for just a few days over #NeurIPS2024. Looking especially for Bioinformatics + Structural Biology skills, MD/Stat Mech skills and/or #deeplearning architecture design + engineering skills
aka.ms/ai4science-r...
11.12.2024 21:20 — 👍 29 🔁 13 💬 0 📌 0
do not assess & compare their methods on fair footing & in a meaningful way
For good or for bad, in the end, BLINDED COMMUNITY CHALLENGES are the only fair way to assess methods
Meaning: the proof is in the pudding. The methods who want to be taken seriously should ...
09.12.2024 03:46 — 👍 8 🔁 2 💬 1 📌 0

Dimensionality reduced embedding space showing that ligand and their targets colocate and genomes self organize.

SPRINT Enables Interpretable and Ultra-Fast Virtual
Screening against Thousands of Proteomes
Andrew T. McNutt ∗
University of Pittsburgh
anm329@pitt.edu
Abhinav K. Adduri∗
CMU, Arc Institute
abhinav.adduri@arcinstitute.org
Caleb N. Ellington∗
Carnegie Mellon University
cellingt@cs.cmu.edu
Monica T. Dayao
Carnegie Mellon University
mdayao@cs.cmu.edu
Eric P. Xing
CMU, MBZUAI, Petuum Inc.
epxing@cs.cmu.edu
Hosein Mohimani
Carnegie Mellon University
hoseinm@cs.cmu.edu
David R. Koes
University of Pittsburgh
dkoes@pitt.edu
We're looking forward to presenting this @workshopmlsb.bsky.social . This work was conceived and led by a group CPCB (@cmupittcompbio.bsky.social) students and learns a co-embedding of proteins and ligands to support ultra fast virtual screening. arxiv.org/pdf/2411.15418
26.11.2024 13:28 — 👍 16 🔁 4 💬 1 📌 2
I teach chemistry to computers
The official Bluesky account of Last Week Tonight with John Oliver. youtube.com/lastweektonight
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Amaro Lab (UC San Diego), PhD student Biochemistry and Molecular Biophysics | Computational Biology
Assis. Prof. @ucsbece Affiliate @SLAClab Stanford Prev @Stanford @Inria @imperialcollege @Polytechnique PI @geometric_intel
http://gi.ece.ucsb.edu, Pilot
International Conference on Learning Representations https://iclr.cc/
RA @ Vanderbilt | Computational Structural Biology
Associate Professor at Columbia Chemistry. PI of a chemical biology lab full of awesome people. Internal conflicts: Chemist or biologist? Kinase or phosphatase?
Lab website: https://shahlab.wixsite.com/home
ORCiD: 0000-0002-1186-0626
Biologist and professor studying evolution of sex & committed to social justice. A rising tide lifts all boats. Senior/Preprint Editor @ Royal Society of London's Proceedings B. Opinions mine and do not represent my employer.
❤️🐌💉
Loves worms, dogs and nothing else. Opinions are my own and rarely reflect those of my employer.
Associate Professor at McMaster University.
Husband, Father, Grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science, Stand Up for Science advisor, Shenanigator, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
https://palermolab.com
Giulia Palermo Lab at the University of California, Riverside. Biophysicist passionate about Science & Art! In love with Nucleic Acids and Computational Science!
CMU-Pitt Computational Biology PhD Candidate
PhD student @ CMU/Pitt Comp Bio
Google Chief Scientist, Gemini Lead. Opinions stated here are my own, not those of Google. Gemini, TensorFlow, MapReduce, Bigtable, Spanner, ML things, ...