We held the first #CANPOP symposium today (cancer population genetics club) @icr.ac.uk. 6 excellent & didactic talks, all applying mathematical #popgen theory to dissect somatic evolutionary dynamics: spanning the full range of pre-cancer, metastasis through to 6000 year old transmissible cancer...
01.12.2025 20:37 — 👍 20 🔁 4 💬 1 📌 0

Epigenetically driven and early immune evasion in colorectal cancer evolution - Nature Genetics
This study nominates immune escape as an early event in colorectal cancer and shows how this can be driven through both genetic and epigenetic changes.
New @natgenet.nature.com paper from the brilliant @eszterlakatos.bsky.social presents evidence that chromatin alterations disrupt antigen presentation & neoantigens in colorectal cancer. Also that immune escape is part of the "Big Bang", at the outset of CRC growth. www.nature.com/articles/s41...
05.11.2025 16:36 — 👍 30 🔁 12 💬 2 📌 3

Big, beautiful trees!!
SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life.
www.biorxiv.org/content/10.1...
14.10.2025 14:37 — 👍 46 🔁 15 💬 1 📌 0

Development Fund
CRUK City of London Centre Development Fund
The CRUK City of London Centre brings together the discovery science of the Crick and the cross-disciplinary, translational and clinical expertise at UCL,...
Huge congratulations to the lead applicants to our Development Fund 2025 on their innovative ideas! 👏 Özgen Deniz; Jonathan Fisher; Paul Grevitt; Jit Sarkar; Adam C Sedgwick; Emmanouil Stylianakis; and @zaccasimo.bsky.social 🎉 Find out more about their projects here: www.colcc.ac.uk/development-...
25.09.2025 11:55 — 👍 3 🔁 1 💬 0 📌 0
Congratulations everyone, outstanding work!
12.09.2025 15:39 — 👍 1 🔁 0 💬 0 📌 0

Fluctuating DNA methylation tracks cancer evolution at clinical scale - Nature
Cancer evolutionary dynamics are quantitatively inferred using a method, EVOFLUx, applied to fluctuating DNA methylation.
Studying cancer evolution needs multi-region or single cell seq for phylogenetics, right? Amazingly (I think!) we found single-sample bulk methylation suffices, via analysis of "fluctuating methylation". In @nature.com today led by brilliant @calumgabbutt.bsky.social www.nature.com/articles/s41...
10.09.2025 15:21 — 👍 91 🔁 39 💬 7 📌 2
🧨Exciting new pre-print 🧨 Introducing ppmSeq - cost effective, scalable dual-strand 🧬🧬 sequencing. Scalability & ultra-low error in one! A game changer for somatic genome discovery science and cfDNA clinical apps. Check out thread for deets!
14.08.2025 18:11 — 👍 22 🔁 5 💬 0 📌 0

UCL – University College London
UCL is consistently ranked as one of the top ten universities in the world (QS World University Rankings 2010-2022) and is No.2 in the UK for research power (Research Excellence Framework 2021).
🚨 We're hiring a sequencing technician in #TRACERx!
Join world-leading teams at @TheCrick & @uclcancer, working at the forefront of cancer research.
Want to apply cutting-edge genomic tech to understand cancer evolution? Apply now!
👉 www.ucl.ac.uk/work-at-ucl/...
#Genomics #CancerResearch
14.07.2025 12:44 — 👍 4 🔁 1 💬 0 📌 1
Not only are you a brilliant PI and an incredible mother, but you’re also a role model for women in science. Your strength and dedication inspire us all!
10.05.2025 05:15 — 👍 10 🔁 1 💬 0 📌 0

I'm participating the ISMB/ECCB 2025 conference, join me!
Register now
I'm attending #ISMBECCB2025 in Liverpool, United Kingdom from July 20-24. Who else is going? Register here and don't miss it! invt.io/1bxbyktm0wq
07.04.2025 20:36 — 👍 1 🔁 0 💬 0 📌 0
Huge congratulations to now PhD @alexsteinresearch.bsky.social and supervisor @benjaminwerner.bsky.social , well deserved! It has been a pleasure to listening to and discussing such a great amount of work and how it all came together
21.03.2025 17:49 — 👍 6 🔁 0 💬 0 📌 0
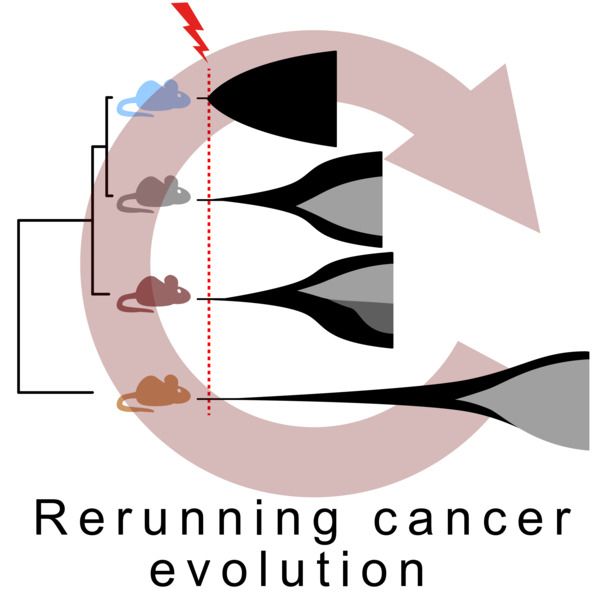
Manuscript logo, phylogenetic tree of mouse strains with different but highly reproducible patterns of cancer evolution. Demonstrated by rerunning cancer evolution in a controlled system.
To what extent is cancer development deterministic and predictable..?
Does the germline genome affect that predictability...?
Preprint: www.biorxiv.org/content/10.1...
15.01.2025 19:31 — 👍 17 🔁 8 💬 2 📌 0
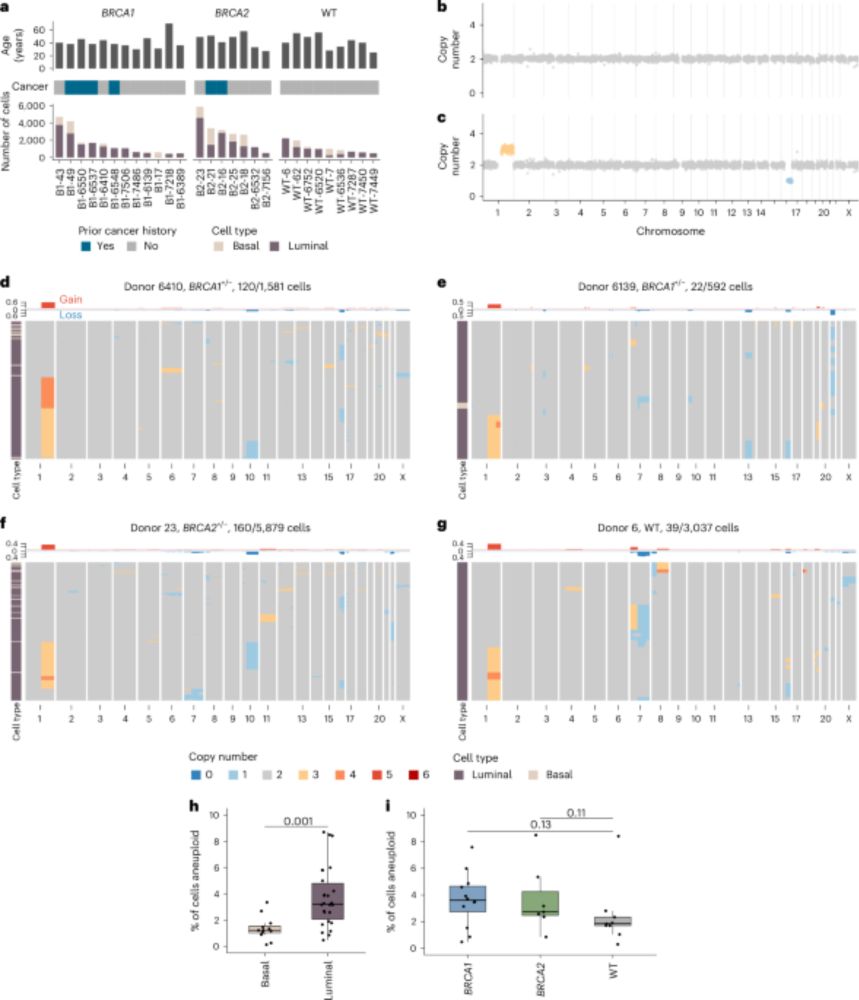
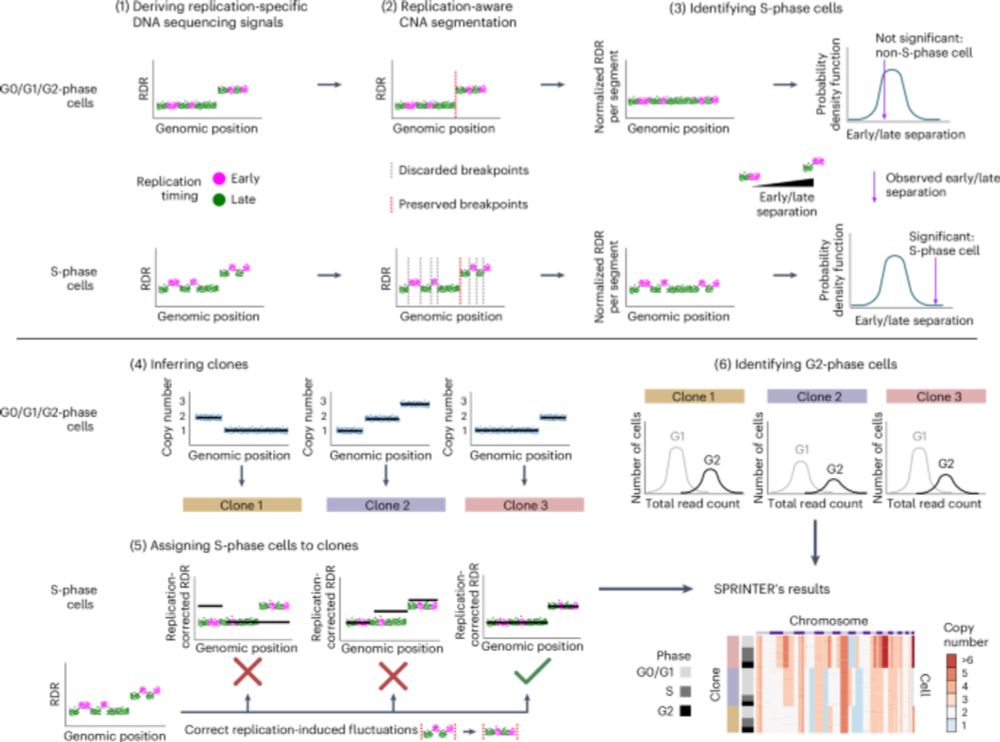
A big moment for our Centre - the first algorithm to identify cells responsible for aggressive tumour growth.
SPRINTER can measure proliferation of distinct clones within the same tumour, using DNA sequencing data. This could facilitate better prediction of cancer progression, informing treatment.
18.12.2024 10:27 — 👍 2 🔁 1 💬 0 📌 0
🚨 Very proud of this commentary led by @rijazaidi.bsky.social reasoning on the impact of methods and experimental design on cancer evolution understanding, motivated by the outstanding work of @TheBoutros, @VanLooLab, @kellrott et al
Original work 👉 doi.org/10.1038/s415...
Our Commentary 👇
16.12.2024 15:47 — 👍 1 🔁 1 💬 1 📌 0
Reposting this list of #cancer #evolution scientists, in case its useful for the many newcomers.
06.12.2024 09:46 — 👍 2 🔁 3 💬 0 📌 0
So proud that our paper introducing SPRINTER to characterise clone proliferation through cancer evolution is out in Nature Genetics!
05.12.2024 09:36 — 👍 2 🔁 1 💬 0 📌 0

Twenty-seven scientists become EMBO Young Investigators – Press releases – EMBO
The group leaders join an international network of nearly 800 life scientists and receive financial support
🎉 Hugely honoured & happy to be selected for the
@embopress.org EMBO Young Investigator Programme 2024!
Exciting opportunity for our group @ccg-ucl.bsky.social & grateful for the support of @uclofficial.bsky.social @uclnews.bsky.social & funders @cancerresearchuk.org
www.embo.org/press-releas...
05.12.2024 12:02 — 👍 6 🔁 0 💬 1 📌 1
We are so grateful to @qianxidu.bsky.social for this outstanding News&Views piece highlighting our recent SPRINTER work and framing it so beautifully within the literature. A truly valuable resource! 🙏
Read it here: 👉 doi.org/10.1038/s415...
🧵 Dive into SPRINTER's work: 👉 bsky.app/profile/zacc...
03.12.2024 19:16 — 👍 2 🔁 0 💬 0 📌 0
What an absolute pleasure to be collaborating with such an incredibly talented team!!
SPRINTER out today in Nature Genetics 🧬
29.11.2024 19:38 — 👍 6 🔁 1 💬 0 📌 0
A great pleasure to work with @zaccasimo.bsky.social and his team on this elegant data and tool.
29.11.2024 19:02 — 👍 5 🔁 1 💬 0 📌 0
Thank you, Trevor!
29.11.2024 18:25 — 👍 0 🔁 0 💬 0 📌 0
Thank you, Wei! 😊
29.11.2024 17:42 — 👍 0 🔁 0 💬 0 📌 0
All of this was really only possible thanks to the support of our funders @cancerresearchuk.org also inc @RosetreesT @bcrfcure.bsky.social @wellcometrust.bsky.social , our teams @uclofficial.bsky.social @thecrick.bsky.social @CRUKLungCentre, and the patients & participants #TRACERx #PEACEautopsy
29.11.2024 13:25 — 👍 0 🔁 0 💬 0 📌 0

This is also the first step and output of my @cancerresearchuk.org Career Development Fellowship, and you can read its overall vision and goal in a recent
@cancerresearchuk.org 's blog post 👇
news.cancerresearchuk.org/2024/02/27/u...
29.11.2024 13:25 — 👍 0 🔁 0 💬 1 📌 0
This was led by stellar @ojlucas.bsky.social and wouldn't have been possible without @charlesswanton.bsky.social @NnennayaKanu cosupervision, @sophie-ward.bsky.social key collaboration, @rijazaidi.bsky.social @abibunkum.bsky.social key contributions and help of many others inc. Mariam and Nicky
29.11.2024 13:25 — 👍 2 🔁 0 💬 1 📌 0
📰Read full story 👉 rdcu.be/d1S6m
🖥️SPRINTER is fully available in GitHub 👉 github.com/zaccaria-lab...
🚆Distributed through Bioconda (with also related container) 👉 bioconda.github.io/recipes/spri...
💾also with a reproducible capsule in CodeOcean👉 doi.org/10.24433/CO.... pic.x.com/zpL2rhbRhl
29.11.2024 13:25 — 👍 0 🔁 0 💬 1 📌 0
Cancer genetics at Imperial College London
- working on #breastcancer and #coloncancer, and a mix and match of cool side projects
Assistant Professor at UBC and BC Cancer Research Institute
Lung cancer | Tumour microenvironment | Spatial biology
https://www.bccrc.ca/dept/io/people/katey-enfield
Biological oceanographer & phytoplankton connoisseur. Forest Lover. Stand up #TEDx person.🧚🏼♀️🦄🔬🌊 Views=mine. @institutrb.bsky.social
@BigelowLab.bsky.social @EMBO_YIN
https://jelenagodrijan.wixsite.com/godi
Group leader @gustaveroussy.fr. Computational Oncology 💻 | Cancer genomics 🧬 | Clonal Hematopoiesis and Leukemia🩸
PhD student; Oxford University & Moffitt Cancer Center; Mathematical Oncology (treatment scheduling and evolutionary models) and Epidemiology (inference and agent-based modeling).
Lecturer in Applied Maths at St Hilda's College, Oxford.
Chief Scientific Officer, CRUK Lung Cancer Centre of Excellence
We study the drivers of cancer evolution from initiation to response to therapy. We are part of the Centre for Cancer Evolution at Barts Cancer Institute and the Francis Crick Institute in London, UK. Posts from Francesca
Associate Professor in Tumour Immunology - UCL Cancer Institute @uclcancer
Honorary Consultant Paediatric Oncology - Great Ormond Street Hospital
Development and clinical translation of engineered T cells for #childhood cancer
@cancergrand NexTGen
MD/PhD Student at Charité | Studying cancer evolution at the Naxerova Lab of HMS Genetics
Professor of Genetics and Pediatrics, UPenn/CHOP🧬. https://orcid.org/0000-0003-2025-5302. Views my own. 🏴🇺🇸
Cancer biologist fascinated by the tumor microenvironment. Postdoc at the
Francis Crick Institute in London.
Senior Bioinformatician @ LifeArc
Metastatic Niche Lab @QMBCI.bsky.social. Studying the role of Tumour Microenvironment in Metastasis formation. @CancerResearchUK.org Fellow. Previously at @Crick.ac.uk, @CRG.eu and @TIGEM.bsky.social
Group Leader at Peter MacCallum Cancer Centre
DoD Ovarian Cancer Academy
Ovarian & Endometrial Cancer | Treatment Resistance
Cancer Genomics | Bioinformatics
Emmy Noether Group Leader @CECAD
Physician-scientist, Haematology & Oncology @UniCologne @UKKoeln
postdoc @ MRC BSU • machine learning for biology • kirkham.co
Genomics, Bioinformatics.
github.com/tobiasrausch