
GitHub - angelolab/toffy: Scripts for interacting with and generating data from the commercial MIBIScope
Scripts for interacting with and generating data from the commercial MIBIScope - angelolab/toffy
We developed a dedicated pipeline for mibi data: github.com/angelolab/to..., but for other data modalities I’m not as familiar what people generally do. Right now there isn’t a good cross platform solution for data normalization, at least not that we’ve found
07.02.2025 01:41 — 👍 1 🔁 0 💬 0 📌 0
Great point. We spent a lot of effort addressing batch effects earlier in our processing pipeline so that SpaceCat wouldn't have to deal with them. In general, I would say the earlier you can address your batch correction issues, the better, but there aren't as many options for spatial data
30.01.2025 00:06 — 👍 3 🔁 0 💬 1 📌 0
Thanks Kieran!
29.01.2025 16:38 — 👍 0 🔁 0 💬 0 📌 0
If you run into any problems getting the codebase to work, have questions about what we found, or want to chat, please don’t hesitate to reach out (/end) bsky.app/profile/noah...
29.01.2025 16:18 — 👍 1 🔁 0 💬 1 📌 0
This wouldn’t have been possible without an amazing team (most of whom have not migrated over to the good place yet!), including Iris, Cami, Seongyeol, Manon, as well as Christina, Marleen and Mike (9/x)
29.01.2025 16:17 — 👍 1 🔁 0 💬 1 📌 0

GitHub - angelolab/SpaceCat: Generate a spatial catalogue from multiplexed imaging data
Generate a spatial catalogue from multiplexed imaging data - angelolab/SpaceCat
This was just a sampling of what we found; for the full details, please check out the paper, as well as our github, where we’ve made all the underlying code open source and available (8/x)
github.com/angelolab/Sp...
29.01.2025 16:17 — 👍 2 🔁 0 💬 1 📌 0
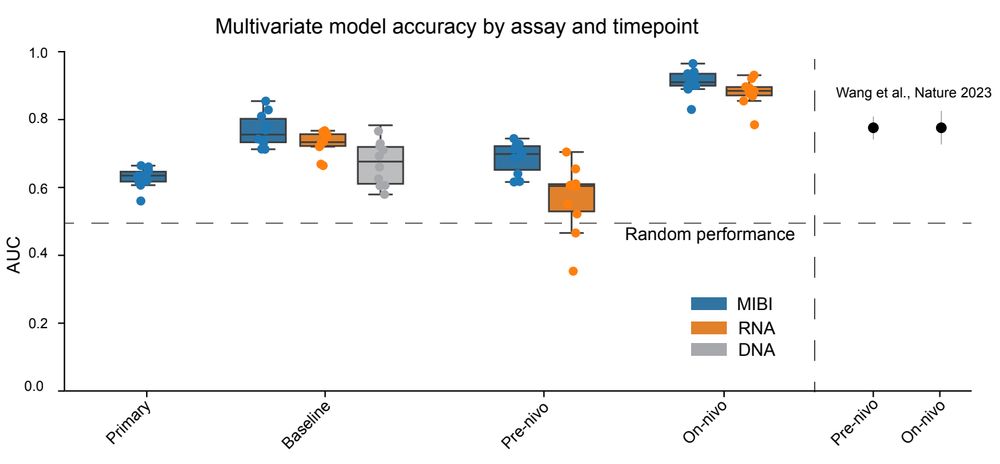
Evaluation of multivariate models trained on different timepoints (x axis) and data types (colors) to predict patient outcome.
Finally, to look at how these features could be combined together, as well as to compare modalities, we built multivariate models to predict outcome from each data type at each timepoint. We found large differences across both assay types and sample timepoints! (7/x)
29.01.2025 16:17 — 👍 0 🔁 0 💬 1 📌 0

The same feature (T / Cancer Ratio) has no association with outcome when looking at the primary tumor, but very strong association with outcome when looking at the on treatment (on-nivo) sample
When we looked at the specific features we defined, we found some that were temporally dependent, with good predictive power at one timepoint but poor predictive power at another timepoint (6/x)
29.01.2025 16:17 — 👍 0 🔁 0 💬 1 📌 0

Volcano plot on the left showing association with outcome for each of the SpaceCat features. Barplot on the right shows the enrichment in top predictive features for those defined within specific regions (compartments) of the tumor
We then tested which of the 800+ features from SpaceCat could predict response to immunotherapy, finding numerous strong predictors. Interestingly, features defined in specific regions of the tumor did an especially good job at predicting outcome (5/x)
29.01.2025 16:17 — 👍 0 🔁 0 💬 1 📌 0

Summary of the types of features that SpaceCat generates, with representative images from four of the categories
To help us make sense of this spatially-resolved data, we built SpaceCat, an algorithm to quantify and summarize the key features from spatial datasets. SpaceCat can be applied to processed imaging data from any multiplexed imaging platform! (4/x)
29.01.2025 16:17 — 👍 0 🔁 0 💬 1 📌 0

Heatmap showing the cell types identified in our study. Each row is a cell type, and each column is a different marker on the antibody panel
We then generated highly multiplexed imaging data using an antibody panel of 37 antibodies. This allowed us to identify 22 cell types across the more than 650 TMA cores we imaged from 117 total patients (3/x)
29.01.2025 16:17 — 👍 0 🔁 0 💬 1 📌 0

Cartoon overview of the samples collected from patients at each timepoint, as well as the number of different modalities (MIBI, DNA, RNA) collected from each.
Our awesome collaborators at NKI put together a unique cohort spanning primary disease, pre-treatment metastases, and on-treatment metastases from triple negative breast cancer patients enrolled in the TONIC clinical trial (2/x)
29.01.2025 16:17 — 👍 2 🔁 0 💬 1 📌 0
I wanted to write briefly about a very pleasant experience we recently had coordinating and collaborating closely on competing publications with 2 other teams. 1/
24.01.2025 19:36 — 👍 114 🔁 23 💬 2 📌 5
Hi Erik, I work on tissue imaging, spatial biology, and cancer research. Could you please add me to the feed? Thanks!https://scholar.google.com/citations?user=ajvnimEAAAAJ&hl=en
24.01.2025 20:27 — 👍 1 🔁 0 💬 0 📌 0
Reposting our Penn Postdoctoral Fellowship in Genetics!
www.med.upenn.edu/apps/my/bpp_...
12.12.2023 16:20 — 👍 1 🔁 1 💬 0 📌 0
Hi all, I just joined! I’m a PhD student at Stanford studying tumor immunology. Excited to try this thing out #HiSciSky #AcademicSky
01.10.2023 19:19 — 👍 20 🔁 3 💬 1 📌 0
MD/PhD Student at Stanford interested in immune signaling, protein engineering, autoimmunity and cancer. PhD with Chris Garcia on a Hertz fellowship.
Computational biology postdoc at Stanford in the Angelo lab. Multiplexed imaging, single-cell bioinformatics, cell fate dynamics :)
Immunologist, Instructor at Stanford University; all things macrophages, spatial tissue biology
Official Bluesky account of the Bendall Lab in the Stanford Department of Pathology and Immunology / Cancer Biology / Stem Cell research and training programs.
Incoming postdoc at Genentech | PhD from Stanford Biomedical Data Science.
Cancer scientist and oncologist, Professor at Stanford and Director of the Stanford Cancer Institute
We are a world leader in genomics research. We apply and explore genomic technologies to advance the understanding of biology and improve health 🧬
https://www.sanger.ac.uk/
Faculty in Generative and Synthetic Genomics at Wellcome Sanger Institute; CTO (part-time) at ExpressionEdits. Engineering DNA in mammalian cells to understand and build.
shendure lab |. krishna.gs.washington.edu
CompBio Scientist, PhD • #spatialomics, #rnabiology, #opensource • Chinese-Malay American
🍱 Creator of github.com/ckmah/bento-tools
⭐️ clarencemah.me
Cross-Institutional Journal Club. We review immunology #preprints and highlight our favourites. Check our website 👉 https://www.preprintclub.com/
Principal Investigator of the Spatial Immunology Unit | Independent Research Scholar | NIH/NIAID Intramural Research Program
Spatially mapping the immune ecosystem of infection and inflammation
(Views are my own)
Assistant Professor @ McMaster University. Studying the impact of inherited DNA variants on tumour evolution.
Postdoc at Stanford Medicine, Curtis Lab. Studying cancer evolution, genotype-to-phenotype relationship, and tumor microenvironments by analyzing various data.
PhD candidate at UChicago. Stem cells, cardiac organoids, gene regulation, single-cell approaches (yes, karyotyping counts).
scientist. shipman lab at gladstone | UCSF.
PI @ Gladstone Institutes & UCSF. Molecular technologies & the genomics / molecular biology / biochemistry of gene regulation. Views here mine & do not represent those of my affiliated institutions.
Assistant Professor at @UCSF_BTS | @HHMINEWS HGF | | Loving membrane proteins every day https://www.wcoyotelab.com










