We have significantly improved the protocol for recording CRE activities and orders with ENGRAM. We also designed new plasmids for easier cloning. Great work with @jennynathans.bsky.social, @troymcdiarmid.bsky.social and @jshendure.bsky.social . Happy recording!!
13.02.2026 19:37 —
👍 9
🔁 5
💬 0
📌 0

Summer Undergraduate Research Experience | HHMI
The Cech Fellows Program is a paid, nine-week summer research experience empowering the next generation of scientific leaders.
Calling all rising juniors & seniors: Interested in biological or biomedical research? Applications for our ’26 Summer Undergraduate Research Experience Program are now open! Nine weeks, hands-on research, & mentorship from some of the nation’s top scientists — learn more: bit.ly/CechFellows
18.11.2025 14:44 —
👍 70
🔁 88
💬 0
📌 12
Our work developing a parts list of promoters and gRNA scaffolds for mammalian genome engineering and molecular recording is now out @natbiotech.nature.com
12.11.2025 18:06 —
👍 24
🔁 10
💬 1
📌 0
🚀 Very excited to share the first major work from my PhD!!
We combined MPRA and CRISPRa in excitatory neurons to test and validate cis-regulation therapies for hundreds of haploinsufficient neurodevelopmental disorder genes. 🧬🔬
www.biorxiv.org/content/10.1...
06.11.2025 23:56 —
👍 18
🔁 5
💬 1
📌 1
Stoked to share our latest work entitled: “Large-scale discovery of neural enhancers for cis-regulation therapies”
shorturl.at/H3Qww
This is an enormous team effort that I had the honour of spearheading with Nick Page and Florence Chardon.
Bluetorial below.
05.11.2025 15:09 —
👍 34
🔁 14
💬 2
📌 3


Thrilled to share I’ve started my lab at Dartmouth’s Geisel School of Medicine! We focus on mapping cellular trajectories & TF networks in development and Mendelian disorders, exploring new therapies. Join us—postdocs, grads, and scientists welcome! sites.dartmouth.edu/qiulab/
04.11.2025 15:10 —
👍 16
🔁 10
💬 2
📌 0
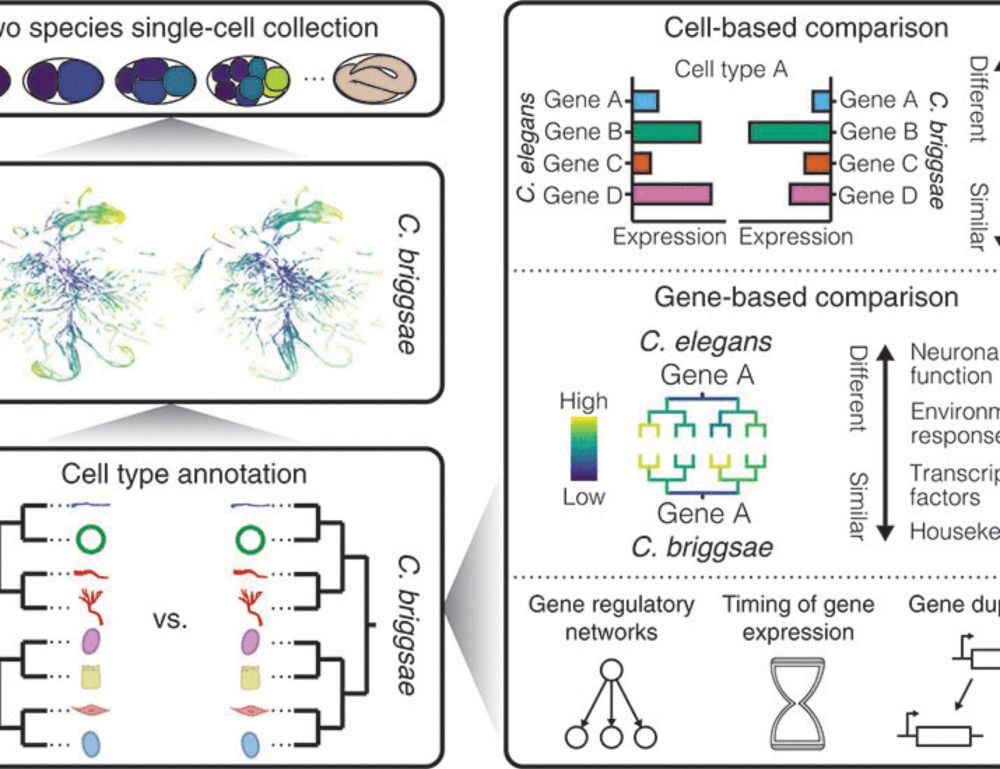
This is one of the coolest figures I've seen in a while
22.10.2025 20:44 —
👍 35
🔁 7
💬 2
📌 0


Super excited about first Shendure/Baker Lab collaboration & preprint on a multiplex sequencing-based strategy for screening de novo proteome editors in mammalian cells. Kudos to the brilliant Chase Suiter (not here) & @greenahn.bsky.social on the work! Preprint here:
www.biorxiv.org/content/10.1...
14.10.2025 18:44 —
👍 97
🔁 33
💬 0
📌 0
There is also no space bar 😢
09.10.2025 11:41 —
👍 4
🔁 0
💬 1
📌 0
Developmental Biology and Disability - the Node
Hopeful monsters. Morphospace. Mutation. Natural variation. Mutagenesis screens. Polymorphism. Deformity. Phenotype. Disease. Adaptation. Anomaly.
Developmental Biology and Disability.
“Developmental biology is fundamentally beautiful. We are no less beautiful for our variation. Instead, perhaps we are more so. Perhaps we are remarkable. Perhaps we are full of wonder.”
Insightful post by Bethan ⬇️👀
thenode.biologists.com/developmenta...
06.10.2025 12:05 —
👍 16
🔁 8
💬 0
📌 1

New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
26.09.2025 18:06 —
👍 54
🔁 20
💬 3
📌 2
Hugely thankful for this 🙏. We will do our best to make the most of it. @erc.europa.eu!
04.09.2025 18:40 —
👍 24
🔁 3
💬 2
📌 0
Thrilled at the news of @10xgenomics.bsky.social ' acquisition of Scale Bio which I co-founded with Frank Steemers (ex-Illumina), @coletrapnell.bsky.social and Garry Nolan . Further thrilled to joining 10X Genomics as an scientific advisor and for the synergies that lie ahead!
07.08.2025 23:32 —
👍 37
🔁 1
💬 3
📌 0


Two new preprints from the @arjunraj.bsky.social on #gastruloids. Both excellent. Exploring the relationship between macroscopic reproducibility v microscopic heterogeneity. Stunning experiments and much to think about
biorxiv.org/content/earl...
biorxiv.org/content/earl...
15.07.2025 13:28 —
👍 44
🔁 16
💬 1
📌 1
🚨 Most variant screens measure growth or abundance. What do they miss? That variants impact a spectrum of protein and cellular phenotypes. Variant in situ sequencing (VIS-seq) finds what’s missing: image cells 🔬 first, decode later, revealing multi-scale phenotypes for thousands of variants.👇
1/9
07.07.2025 02:44 —
👍 48
🔁 18
💬 2
📌 0
One thing that really bothers me with the new "virtual cell" terminology is that it is currently largely focused on a very narrow definition of models that can predict effects of trans perturbations (gene dosage, drugs etc) on gene expression. 1/
28.06.2025 10:38 —
👍 104
🔁 29
💬 1
📌 0
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
13.06.2025 04:30 —
👍 165
🔁 50
💬 11
📌 10

A multi-kingdom genetic barcoding system for precise clone isolation - Nature Biotechnology
A barcoded CRISPR base editing system isolates target clones from complex mammalian, yeast and bacterial populations.
CloneSelect published in Nature Biotechnology
@natbiotech.nature.com. This retrospective clone isolation method using CRISPR base editors is a powerful tool in broad biology. A history of Soh in the Yachie lab. www.nature.com/articles/s41...
21.05.2025 13:56 —
👍 29
🔁 13
💬 3
📌 0

Massively parallel jumping assay decodes Alu retrotransposition activity - Nature Communications
Here, the authors develop a high-throughput assay to measure the jumping potential of thousands of transposons in parallel.
Massively parallel jumping assay (MPJA) enables to test the jumping potential of thousands of transposons. Analysis of >160,000 Alu haplotypes identified transposition-asssociated domains. Amazing work by Navneet Matharu, Jingjing Zhao, Martin Kircher and many others.
www.nature.com/articles/s41...
09.05.2025 03:50 —
👍 77
🔁 35
💬 1
📌 2