The result is a computationally cheap algorithm for connectivity at micro- meso- and macro-scale and a new view on the structure of connectivity. First diving into biological detail we emerged with a simplified model. Validated against the amazing MICrONS data.
25.08.2025 10:08 — 👍 0 🔁 0 💬 0 📌 0
We explain how morphologies work in terms of their constraints on connectivity, why these constraints are stronger than anticipated, and why they provide structure that other models don’t. Understanding this allowed us to capture all of this even without using morphologies.
25.08.2025 10:08 — 👍 0 🔁 0 💬 1 📌 0
How are we to understand the structure of synaptic connectivity at cellular resolution? Some say that predictions from neuronal morphologies can work well. Others point out that this provides limited insights into underlying mechanisms and is computationally too expensive.
25.08.2025 10:08 — 👍 0 🔁 0 💬 1 📌 0
Anyone at CNS*2025 in Florence? I hope to see you on Wednesday in our workshop: Bridging Complexity and Abstraction: Large-Scale Mechanistic Models of Brain Circuits from Biophysically Detailed to Simplified Representations.
07.07.2025 09:00 — 👍 2 🔁 0 💬 0 📌 0

Potentially even for inter-regional connectivity. Hope it will help with our understanding of neuronal connectivity. Readily usable for example in models and simulations of neuronal networks.
28.05.2025 14:13 — 👍 0 🔁 0 💬 0 📌 0

Much has been written about the structure and heterogeneity of local neuronal circuitry that is not captured by controls. We built a surprisingly simple graph based model and show that it recreates the complexity characterized in biological neuronal networks at different scales.
28.05.2025 14:13 — 👍 0 🔁 0 💬 1 📌 0
I have embraced the complexity of the brain and data for a long time, and believe that approach is required. But that does not have to be overly centralized. Can be decentralized and incremental. Just focus on improving existing resources instead of reinventing the wheel.
28.03.2025 13:15 — 👍 1 🔁 0 💬 0 📌 0
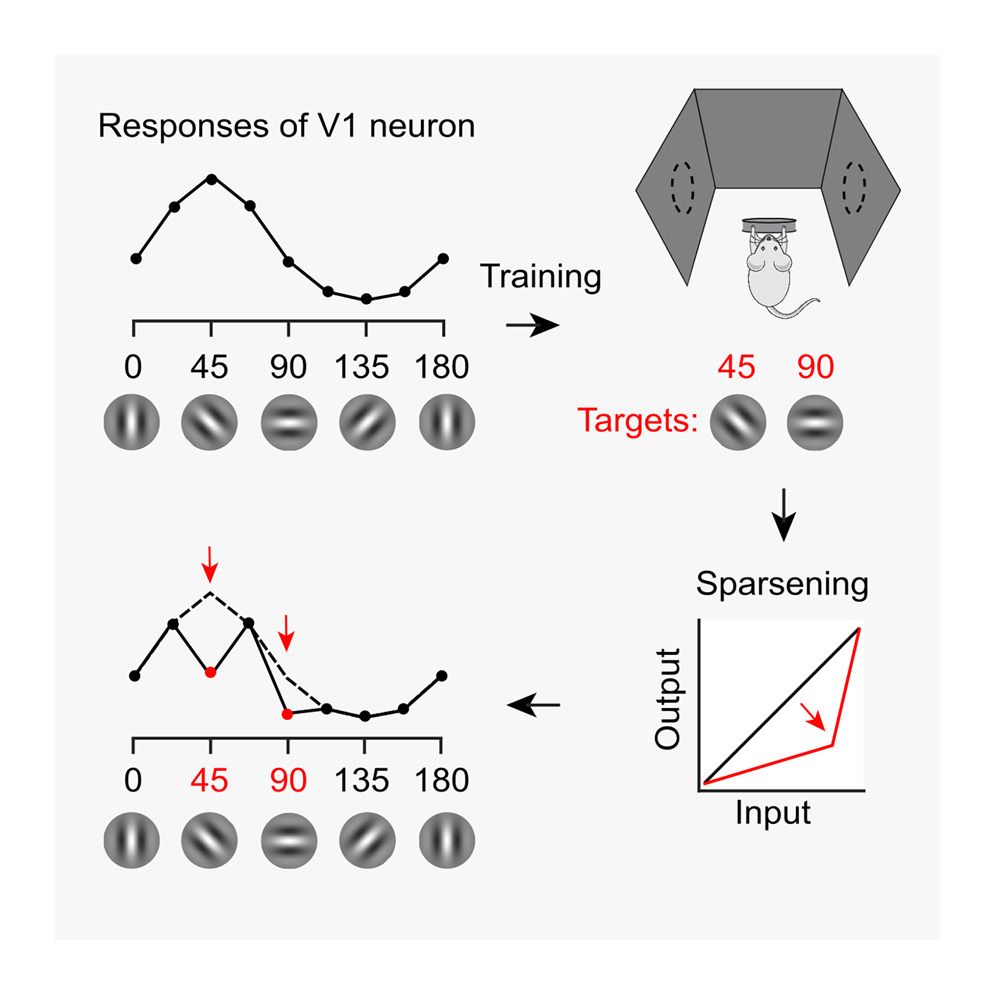
It's finally out!
Visual experience orthogonalizes visual cortical responses
Training in a visual task changes V1 tuning curves in odd ways. This effect is explained by a simple convex transformation. It orthogonalizes the population, making it easier to decode.
10.1016/j.celrep.2025.115235
02.02.2025 09:59 — 👍 153 🔁 44 💬 5 📌 2
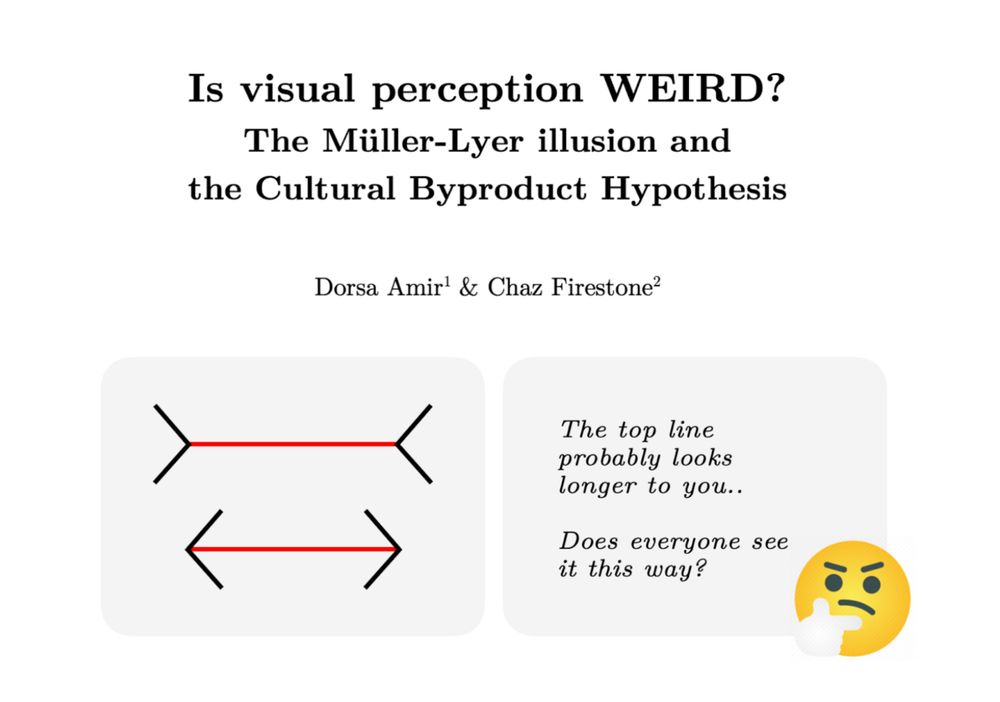
Does the culture you grow up in shape the way you see the world? In a new Psych Review paper, @chazfirestone.bsky.social & I tackle this centuries-old question using the Müller-Lyer illusion as a case study. Come think through one of history's mysteries with us🧵(1/13):
25.01.2025 22:05 — 👍 1095 🔁 423 💬 33 📌 78

Leslie Ungerleider (1946 - 2020) was an extraordinary, pioneering neuroscientist. The latest issue of @jocn.bsky.social honors her legacy with articles authored by former colleagues & trainees that highlight critical aspects of her work and its influence https://buff.ly/4156UKw #cogsci #neuroscience
04.12.2024 00:40 — 👍 74 🔁 29 💬 1 📌 2
Maybe I read too much into the two sentences, but it seems to imply something like: You plug Bernoulli's principle into a computer, then optimize for agent mobility and end up with a weird looking quadcopter. Conclusion: Clearly birds are flying using four rotors.
18.12.2024 09:51 — 👍 1 🔁 0 💬 0 📌 0
petition to change the word describing ChatGPT's mistakes from 'hallucinations' to 'confabulations'
A hallucination is a false subjective sensory experience. ChatGPT doesn't have experiences!
It's just making up plausible-sounding bs, covering knowledge gaps. That's confabulation
11.12.2024 09:47 — 👍 286 🔁 57 💬 25 📌 12

Michael W. Reimann
Group Leader @ Blue Brain, EPFL, Switzerland - Cited by 3,298 - cortical circuits - modeling - connectomics - neural codes
Hello Monica @pkpd-babe.bsky.social . Could you please add me to the science feed? I am a neuroscientist working on connectomics with simulation-based methods. Scholar profile: scholar.google.com/citations?us...
06.12.2024 09:23 — 👍 0 🔁 0 💬 0 📌 0
🤔 Yeah that makes a lot of sense to me.
I like this way of thinking in the context of constraints, in this case: the imperative to conserve energy. Needed to understand the brain, in my opinion.
03.12.2024 17:13 — 👍 1 🔁 0 💬 0 📌 0
I would ask to what degree the current parcellation into brain regions works for describing the rules of the mapped connections. Can we improve it further?
03.12.2024 16:19 — 👍 0 🔁 0 💬 0 📌 0
But properly manipulating connectivity in detailed models is harder than it sounds. Changing one aspect while keeping others fixed is tricky due to complex dependencies. Our tool helps with this. We show examples and results of how this plays out in practice.
03.12.2024 14:42 — 👍 0 🔁 0 💬 0 📌 0
Ultimately, this can only be tested through manipulations that are currently only possible in simulation. Due to the wealth of information EM gives us about connections (dendritic locations, sizes of synapses, types of participating neurons) we would do this in very detailed models.
03.12.2024 14:42 — 👍 0 🔁 0 💬 1 📌 0

The motivation for the work came from ongoing and published electron microscopic connectome reconstructions that were said to solve the connectomics of brain microcircuitry. But even knowing all connections, one can describe any number of rules and trends - which ones are functionally meaningful?
03.12.2024 14:42 — 👍 1 🔁 0 💬 1 📌 0
Brain region models and papers | Linktree
Models of rodent hippocampus and somatosensory regions - and papers using them
Our SSCx model (linktr.ee/BlueBrainPjt) provided a valuable null model to compare findings against: It has the excitatory “fan-out tracks”, but not the specific inhibition. Clearly: Inhibition is computationally powerful and should be studied more! (3/3)
19.11.2024 09:32 — 👍 1 🔁 0 💬 0 📌 0

A schematic that describes the main excitatory + inhibitory motif encountered in the data.
We found that the higher-order structure of excitation contains specific motifs that form a backbone of connectivity in a fan-out structure. Inhibitory connectivity is structured by this, providing specific competition between the motifs. Disinhibition in turn targets these neurons (2/3)
19.11.2024 09:32 — 👍 1 🔁 0 💬 1 📌 0

Specific inhibition and disinhibition in the higher-order structure of a cortical connectome
Abstract. Neurons are thought to act as parts of assemblies with strong internal excitatory connectivity. Conversely, inhibition is often reduced to blanke
We are happy to announce that our latest connectomics work has been published by Cerebral Cortex. We have analyzed one of our favorite open datasets: MICrONS cortical mm^3. In this EM dataset we found new rules for neuronal connectivity & describe them (doi.org/10.1093/cerc...) (1/3)
19.11.2024 09:32 — 👍 2 🔁 0 💬 1 📌 0
My favorite sentence from the paper: "Moreover, ANNs usually disregard the fact that biological networks are spatially
embedded networks running under constraints, a feature we discussed earlier as a potential key to understanding
neural network organization."
Could not agree more!
17.11.2024 08:00 — 👍 3 🔁 0 💬 0 📌 0
Circadian Sleep Disorders Advocate. Raising awareness for Non24 in sighted people. #DSPD mom of a #Non24 adult. Former board member at https://csd-n.org.
Cover photo by Daniel Kordan.
[Yes, I follow a lot of accounts! Curious human, not a bot].
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Neuroscience at the speed of thought
www.openbraininstitute.org/
Open source privacy and security focused mobile OS with Android app compatibility.
https://grapheneos.org/
Assistant prof at Baylor College of Medicine. Sensory circuits & brain state modulation. Imaging, patching, and pupils. Kiddo dad. Opinions are my own. (he/him)
Assistant Investigator at Allen Institute for Brain Science. Formerly Janelia, Universität Zürich, and U Mich Physics.
Building bottom-up insight into the brain from synaptic resolution connectomics and making computational tools to help you do that too.
Understanding life. Advancing health.
Dedicated to helping neuroscientists stay current and build connections. Subscribe to receive the latest news and perspectives on neuroscience: www.thetransmitter.org/newsletters/
Luiz Pessoa, University of Maryland, College Park
Neuroscientist interested in cognitive-emotional brain
Author of The Entangled Brain, MIT Press, 2022
Author of The Cogitive-Emotional Brain, MIT Press, 2013
Neuroscience & Philosophy Salon (YouTube)
Clinical psychologist and researcher investigating anxiety disorders, Acute PTSD, dissociations, OCD, Affective Neuroscience, neuropsychoanalysis, application of nonlinear dynamical systems to psychology
https://montgomerycountypsychologist.com
associate prof at university of minnesota | masonic institute for the developing brain | brain networks & behavior lab PI | views my own | oberlin to iu to penn to iu to umn | https://www.brainnetworkslab.com/
Computational Neuroscience PhD Student
Assistant Professor at the University of Newcastle | Fulbright Scholar | MSCA Fellow | Lister Institute Prize Fellow | BoD of the ALBA Network | Chair of IBRO ECC | OIST TSVP Scholar | Cosyne DEIA | Passionate about Comp Neuro, NeuroAI and enhancing DEI.
ELLIS & IMPRS-IS PhD Student at the University of Tübingen.
Excited about uncertainty quantification, weight spaces, and deep learning theory.
Scientist, Open Brain Institute, Lausanne @openbraininst.bsky.social
Past: Postdoc @ EPFL - Blue Brain project,
PhD @ IIT Bombay
https://orcid.org/0000-0002-7104-4604














