
LFQ Benchmark Dataset - Generation Beta: Assessing Modern Proteomics Instruments and Acquisition Workflows with High-Throughput LC Gradients www.biorxiv.org/cont...
---
#proteomics #prot-preprint
@robbedevr.bsky.social
PhD student in computational proteomics

LFQ Benchmark Dataset - Generation Beta: Assessing Modern Proteomics Instruments and Acquisition Workflows with High-Throughput LC Gradients www.biorxiv.org/cont...
---
#proteomics #prot-preprint
Happy to have this one finally out. Since the first generation was very popular in the proteomics community, we decided to work on a second generation including the latest innovations in LC-MS. Very grateful to all collaborators and a special shoutout to @robbedevr.bsky.social from @compomics.com
03.02.2026 09:19 — 👍 7 🔁 2 💬 0 📌 0LFQ Benchmark Dataset - Generation Beta: Assessing Modern Proteomics Instruments and Acquisition Workflows with High-Throughput LC Gradients https://www.biorxiv.org/content/10.64898/2026.01.29.702266v1
02.02.2026 20:47 — 👍 2 🔁 1 💬 0 📌 0
We are happy to share that the preprint for Proteobench is available now on biorxiv!
Check it out here: www.biorxiv.org/content/10.6...
Congratulations and thanks to everyone involved for the great effort!

Exited to share our latest work! Out now in @natcomms.nature.com
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
pubs.acs.org/doi/10.1021/... IM2Deep might win best algorithm name of 2025
15.07.2025 19:59 — 👍 6 🔁 1 💬 1 📌 0
Conference stage with Marie Locard-Paulet, the chairs, and the opening slide in the background.
Marie Locard-Paulet presenting the EuBIC-MS #ProteoBench project at the #EuPA2025 conference.
👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking
MLMarker is live! This ML-tool predicts tissue similarity and uncovers biomarkers from your proteomics data. It was trained on public data of healthy human tissues.
Preprint & app: www.biorxiv.org/content/10.1...
Let's chat at #EuPA2025 - Award session (Wednesday) & poster session (Thursday)!
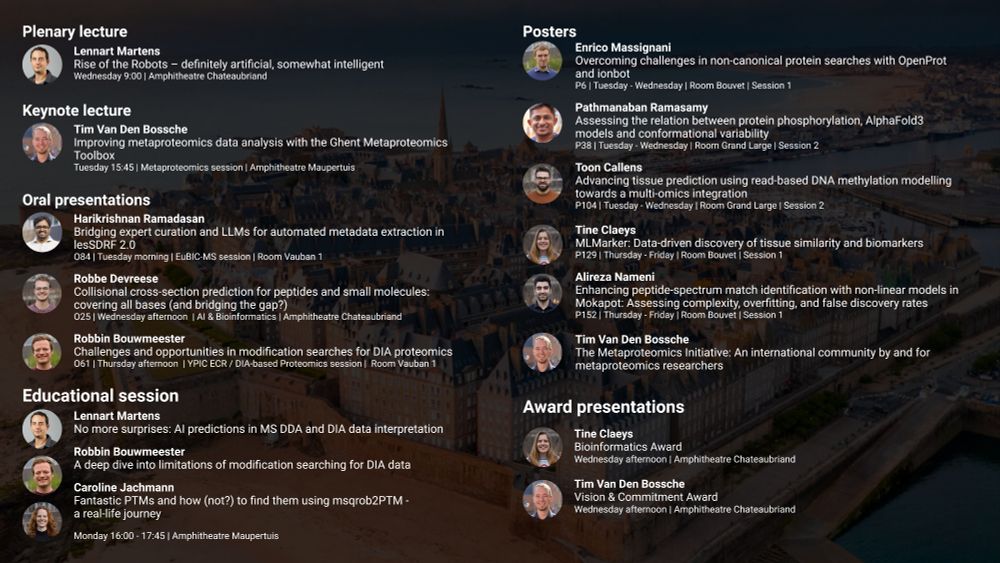
Plenary talk Lennart Martens Rise of the Robots – definitely artificial, somewhat intelligent Keynote lecture Tim Van Den Bossche Improving metaproteomics data analysis with the Ghent Metaproteomics Toolbox Oral presentations Harikrishnan Ramadasan Bridging expert curation and LLMs for automated metadata extraction in lesSDRF 2.0 Robbe Devreese Collisional cross-section prediction for peptides and small molecules: covering all bases (and bridging the gap?) Robbin Bouwmeester Challenges and opportunities in modification searches for DIA proteomics Educational session Lennart Martens No more surprises: AI predictions in MS DDA and DIA data interpretation Robbin Bouwmeester A deep dive into limitations of modification searching for DIA data Caroline Jachmann Fantastic PTMs and how (not?) to find them using msqrob2PTM - a real-life journey Poster presentations Enrico Massignani Overcoming challenges in non-canonical protein searches with OpenProt and ionbot Pathmanaban Ramasamy Assessing the relation between protein phosphorylation, AlphaFold3 models and conformational variability Toon Callens Advancing tissue prediction using read-based DNA methylation modelling towards a multi-omics integration Tine Claeys MLMarker: Data-driven discovery of tissue similarity and biomarkers Alireza Nameni Enhancing peptide-spectrum match identification with non-linear models in Mokapot: Assessing complexity, overfitting, and false discovery rates Tim Van Den Bossche The Metaproteomics Initiative: An international community by and for metaproteomics researchers Award presentations Tine Claeys Bioinformatics Award Tim Van Den Bossche Vision & Commitment Award
From PTMs to proteins, from metadata to metaproteomics. CompOmics has got you covered at #EuPA2025!
16.06.2025 08:05 — 👍 22 🔁 8 💬 0 📌 0
EuBIC-MS session at EuPA 2025 Tuesday June 17, 10:15 - 12:00, Room Vauban 1 Ralf Gabriels Introducing EuBIC-MS 10:15 - 10:30 Julian Uszkoreit O85 - What can we gain - a comparison of common search engines and post-processing methods 10:30 - 10:45 Yannic Chen O84 - Benchmarking Database Search Engines for DDA-based Immunopeptidomics 10:45 - 11:00 Harikrishnan Ramadasan O86 - Bridging expert curation and LLMs for automated metadata extraction in lesSDRF 2.0 11:00 - 11:15 Interactive discussions on open challenges in computational proteomics 11:15 - 12:00
Join us in Saint-Malo for the #EuBIC-MS session at #EuPA2025!
After a short introduction, we have three exciting talks lined up, as well as an interactive discussion on open issues in computational proteomics.
@eupaproteomics.bsky.social @uszkoreitju.bsky.social @harirmds.bsky.social

New in DeepLC! Ability to deal with wild, weird, and wobbly LC setups or peptide modifications. This ability is possible with transfer learning; where only a minimal amount of training peptides are needed for accurate retention time predictions.
www.biorxiv.org/content/10.1...

BEFORE THE BATHROOM SCALE WAS INVENTED, THE ONLY WAY TO WEIGH PEOPLE WAS MASS SPECTROMETRY.
Love today’s #xkcd comic on
#MassSpec
🤣




What an amazing team, this #proteobench people. It was very inspiring to join, support and push this Proteobench project forward representing Core4life during the Proteobench hackaton this week. Thank you to Institut Pasteur, for hosting the joint adventure!
23.05.2025 19:39 — 👍 3 🔁 1 💬 0 📌 0
🚀 New preprint alert! We've improved IM2Deep for accurate peptide collisional cross-section (CCS) prediction, even for peptides exhibiting multiple conformations in the gas phase! 🎯
Check it out here:
www.biorxiv.org/content/10.1...

Collisional cross-section prediction for multiconformational peptide ions with IM2Deep www.biorxiv.org/cont...
---
#proteomics #prot-preprint
What excites me most is that it introduces the first ML-based solution for peptide multiconformers. But that’s not all! We also demonstrate a substantial performance boost for uniconforming peptides.
Our findings are clear: multiconformer peptides cannot be overlooked when predicting CCS!

🚀 New preprint alert! We've improved IM2Deep for accurate peptide collisional cross-section (CCS) prediction, even for peptides exhibiting multiple conformations in the gas phase! 🎯
Check it out here:
www.biorxiv.org/content/10.1...

Seems reasonable to dedicate my first Bluesky post to the following:
Our latest research, TIMS²Rescore, is now published in Journal of Proteome Research! 🎉
Read it here: pubs.acs.org/doi/full/10....
A huge thanks to all our collaborators for making this happen!

Caroline Jachmann presenting her work in front of the BePAc 2024 audience

Robbe Devreese presenting his work at BePAc 2024
Today, @robbedevr.bsky.social and @carojachmann.bsky.social presented their work on #IM2Deep and #ProteoBench at #BePAc2024.
Learn more at doi.org/10.1101/2024... and proteobench.readthedocs.io.