I didn’t know this was already existing
15.12.2025 18:48 — 👍 0 🔁 0 💬 0 📌 0@jonasscheid.bsky.social
#Bioinformatics PhD Student currently focusing on #immunopeptidomics pipeline development @nf-co.re 🧬👨💻
@jonasscheid.bsky.social
#Bioinformatics PhD Student currently focusing on #immunopeptidomics pipeline development @nf-co.re 🧬👨💻
I didn’t know this was already existing
15.12.2025 18:48 — 👍 0 🔁 0 💬 0 📌 0@pride-ebi.bsky.social
15.12.2025 18:47 — 👍 1 🔁 0 💬 0 📌 0Nice! I just mentioned that this feature would be great to have in the PSI-AI meeting. There we have it! I‘ll try it soon thanks!
15.12.2025 07:07 — 👍 0 🔁 0 💬 1 📌 0MCP-Support
14.12.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0
Pipeline release! nf-core/epitopeprediction v3.1.0 - 3.1.0 - Lustnau!
Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/3.1.0
Excited to share our new paper “MHCquant2 refines immunopeptidomics tumor antigen discovery” in Genome Biology!
An open-source @nf-co.re workflow using OpenMS, DeepLC & MS²PIP boosts peptide ID & sensitivity. Scalable, reproducible & ready for large-scale immunopeptidomics.
👉 rdcu.be/eHFTZ

There is such a need to harmonize proteomics data, it’s not even funny anymore. pubs.acs.org/doi/full/10....
06.10.2025 07:30 — 👍 0 🔁 0 💬 0 📌 0Pipeline release! nf-core/hlatyping v2.1.0 - 2.1.0 - Chewbacca!
Please see the changelog: https://github.com/nf-core/hlatyping/releases/tag/2.1.0
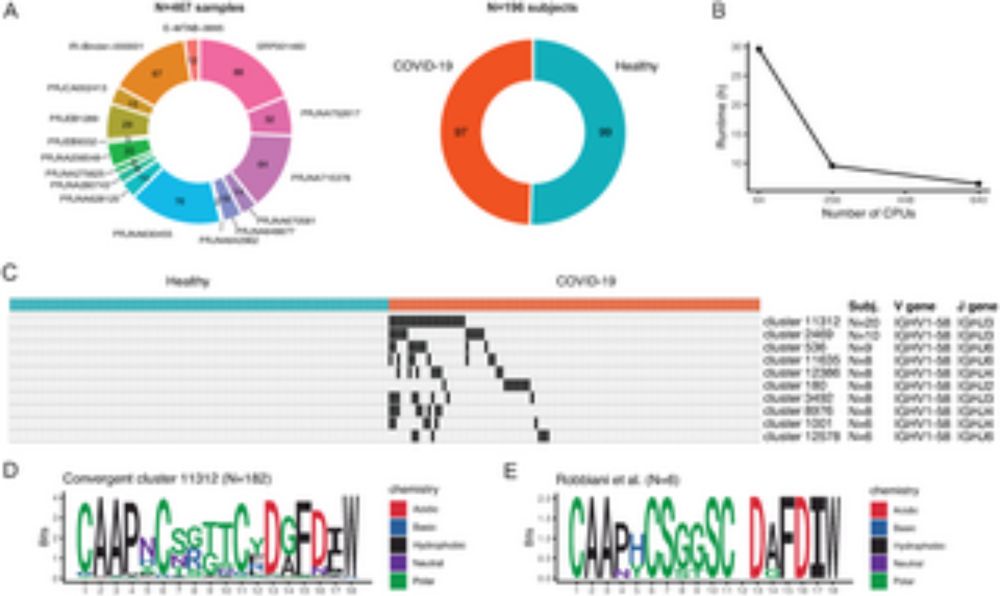
Do you analyze large BCR and TCR sequencing datasets?
Answer our survey for improving nf-core/airrflow, a Nextflow pipeline to analyze bulk and single-cell AIRRseq data (doi.org/10.1371/jour...). We’re interested in your opinion on useful new features!
yalesurvey.ca1.qualtrics.com/jfe/form/SV_...
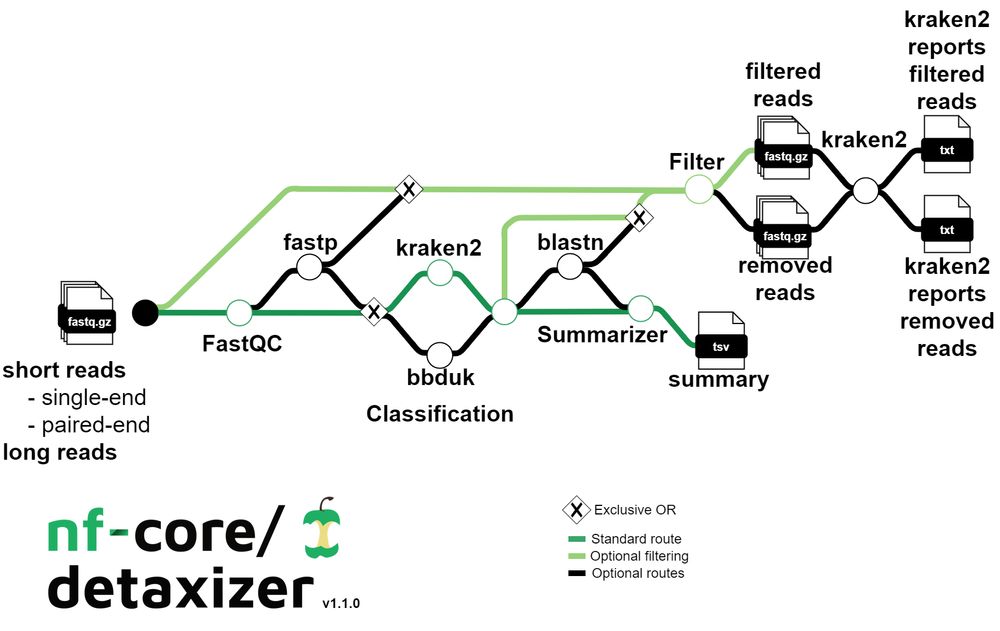
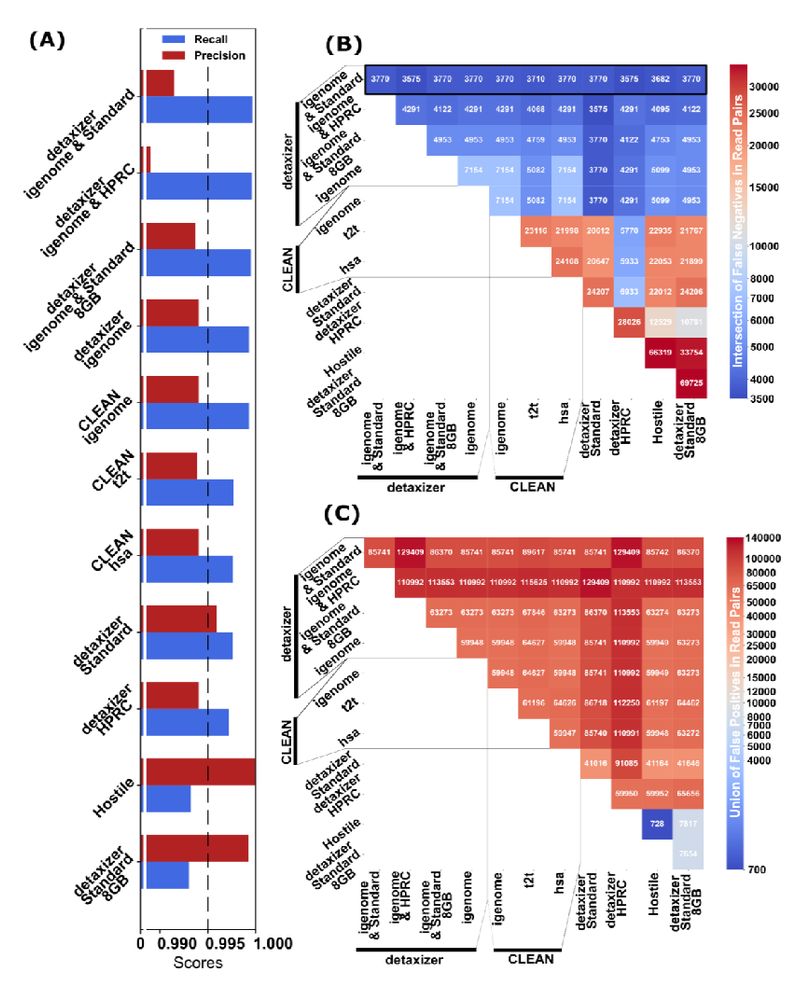
nf-core/detaxizer nf-co.re/detaxizer: A Benchmarking Study for Decontamination from Human Sequences www.biorxiv.org/content/10.1... 🧬🖥️🧪 nextflow github.com/nf-core/deta...
31.03.2025 10:01 — 👍 12 🔁 6 💬 0 📌 0
Interested in contributing to @nf-co.re or improving your nf-core pipeline? Register and join the #nf-core hackathon local hub in Tübingen on March 24th-27th if you live nearby! We have snacks and stickers!
nf-co.re/events/2025/...
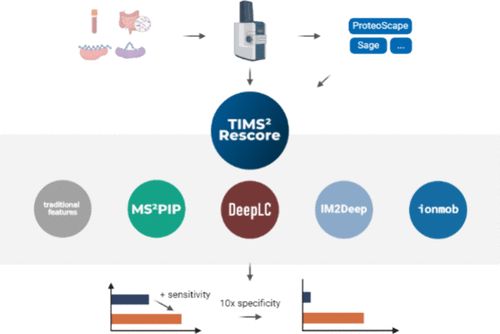
TIMS2Rescore: A Data Dependent Acquisition-Parallel Accumulation and Serial Fragmentation-Optimized Data-Driven Rescoring Pipeline Based on MS2Rescore pubs.acs.org/doi/10....
---
#proteomics #prot-paper
We are #hiring! 🚀 Join our #bioinformatics team in a truly #interdisciplinary environment driving #immunopeptidomics analyses and pipeline development with direct impact on advancing patient treatment! Check out the full job description: jobs.medizin.uni-tuebingen.de/Job/5775/Bio...
04.02.2025 06:43 — 👍 0 🔁 1 💬 0 📌 0
Audience in lecture hall

Pavel Sinitcyn presenting

Katerina Nastou presenting

Will Fondrie presenting
The EUBIC-MS Developers Meeting 2025 in Brixen is off to a great start with keynotes by Pavel Sinitcyn, Katerina Nastou, and @willfondrie.com.
#EuBIC2025
Can’t stress enough about having some code repository (GitHub etc) especially if you are early career and/or looking for a job. If the selection criteria asks for scientific programming skills, everyone claims they can code and use good practice. Show some evidence by providing a link to GitHub etc
01.02.2025 09:13 — 👍 41 🔁 15 💬 2 📌 0Great Stuff! Thanks for the Thread. Is synchro-PASEF now built-in the Bruker software, or is a Special plugin needed?
28.01.2025 13:40 — 👍 0 🔁 0 💬 1 📌 0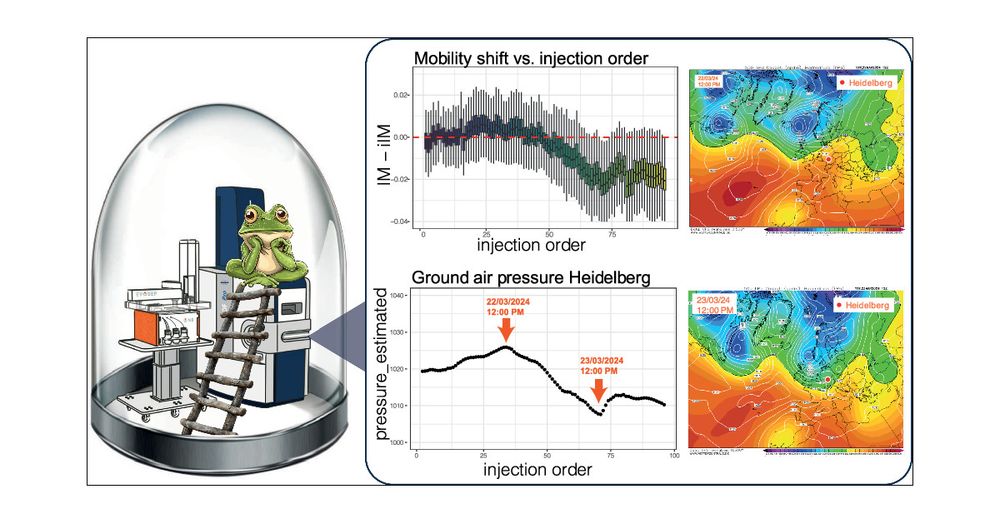
Nice and short technical paper about how air pressure can change ion mobility shifts and peptide quantification! #proteomics
pubs.acs.org/doi/abs/10.1...
I was forced to do 3h pure copy&pasting excel lists as a student assistant and got sick of it quickly
14.01.2025 22:38 — 👍 0 🔁 0 💬 0 📌 0
Phase I trial update: Our warehouse-based cancer peptide vaccine for CLL showed promising safety but limited T cell responses.
Next: Follow-up trial with improved adjuvants & BTK inhibitors underway. Stay tuned! #CancerResearch #Immunopeptidomics #Immunotherapy
🔗 doi.org/10.3389/fimm...
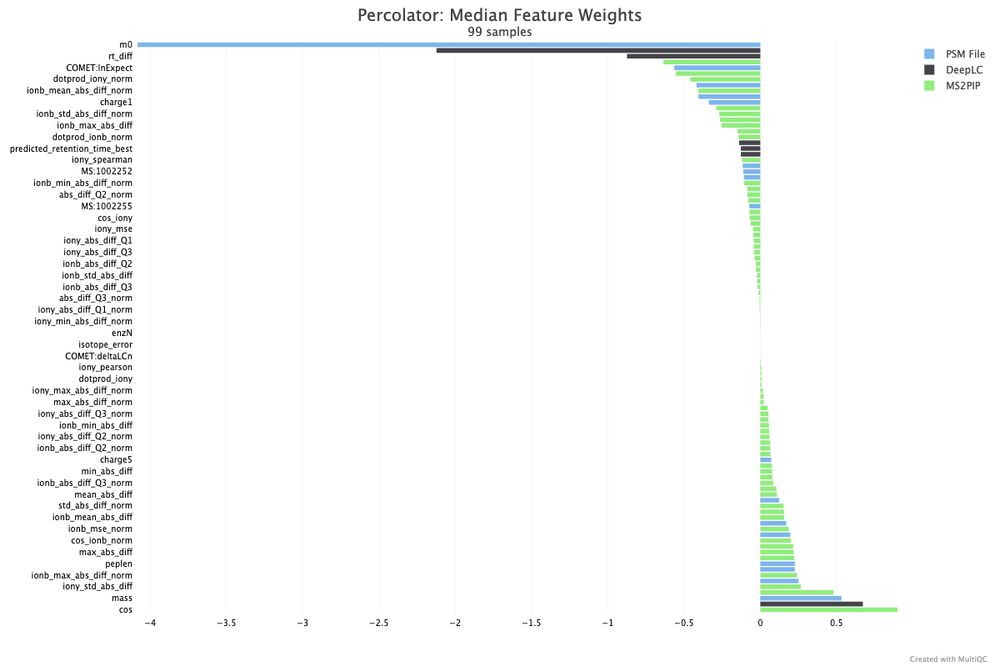
Nice tiny Xmas gift in the new release 🎄. Percolator feature weights can be simply plotted now with multiQC!
22.12.2024 11:50 — 👍 3 🔁 1 💬 0 📌 0
Leveraging the Human Panproteome to Enhance Peptide and Protein Identification in Proteomics and Metaproteomics www.biorxiv.org/cont...
---
#proteomics #prot-preprint
Nice work! I had a hard time using some of these tools with #immunopeptidomics data some time ago. Do you know/recommend a tool here that is decent in analyzing this data?
23.11.2024 08:46 — 👍 0 🔁 0 💬 1 📌 0Happy to announce MS²Rescore v3.0, our much-improved platform for #MachineLearning driven rescoring of #Proteomics search results.
A big thanks to Louise Buur and Viktoria Dorfer for the great collaboration!
Open access full text: biblio.ugent.be/publication/...
Project: github.com/compomics/ms...
Pipeline release! nf-core/epitopeprediction v2.3.0 - 2.3.0 - Oesterberg!
Please see the changelog: https://github.com/nf-core/epitopeprediction/releases/tag/2.3.0