Come learn the tricks of giving a BAD talk: begin the process by organizing your slides - the more, the better, try to impress, run over your time, embrace distractions, and treat questions as threats!
Join us tomorrow at Weill Cornell for this month's Postdoc Night Science NYC!
14.10.2025 12:28 — 👍 25 🔁 7 💬 2 📌 3
Dear Postdocs of New York: Next Wednesday, we’re closing out an amazing year of NYC Postdoc Night Science Club sessions at our very own @weillcornell.bsky.social !
If you’re interested in giving engaging, memorable talks, you cannot miss this one.
Details below, hope to see you all there!
07.10.2025 22:22 — 👍 5 🔁 1 💬 0 📌 0
Great job @jeffvierstra.bsky.social ! Could you recommend a list of QC metrics to check for scATAC-seq datasets, with examples of good and bad quality? and do you have examples of papers that did a great job with the QC? thanks in advance :)
02.09.2025 10:11 — 👍 0 🔁 0 💬 0 📌 0
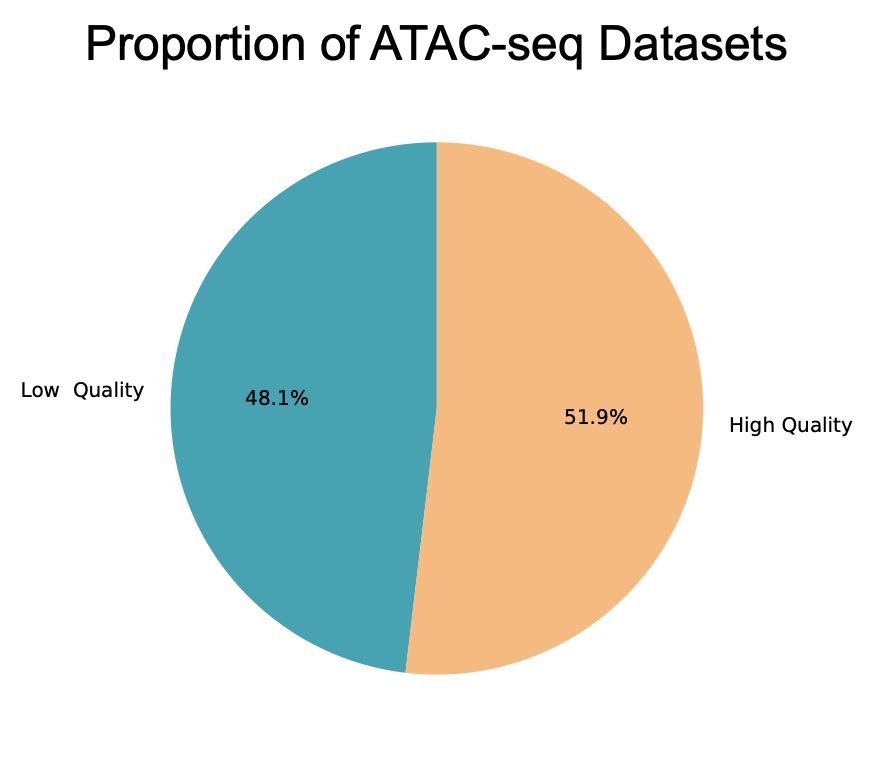
PSA: We just finished processing nearly all public ATAC-seq datasets from SRA (about 22,000 datasets). (Not?) Surprisingly, we had to throw-out nearly ~50% because they were low-quality (low signal-to-noise, duplicate rate, etc.). Check quality before analysis (TSS-enrichment is not sufficient!).
13.05.2025 19:42 — 👍 51 🔁 17 💬 7 📌 2
RAAAAAAHHH!!
20.06.2025 23:24 — 👍 0 🔁 0 💬 0 📌 0
Dear NYC postdocs: the Postdoc Night Science is coming to our very own New York Genome Center on June 2, 5:30-7:30 pm!
Let's dive into the art of asking the right questions, then network with fellow postdocs & find new science buddies 😊
Register below!
29.05.2025 02:28 — 👍 3 🔁 1 💬 0 📌 2

NYC Postdocs! Join us for Night Science at the New York Genome Center on June 2nd for a discussion on the creative process of finding novel questions. This time we hit the bar after! Register: docs.google.com/forms/d/1s9W...
@stearnslab.bsky.social @kelseymonson.bsky.social @rmassonix.bsky.social
14.05.2025 21:30 — 👍 21 🔁 9 💬 1 📌 4
... which may impact downstream analysis such as GRN modeling.
18.03.2025 22:43 — 👍 2 🔁 0 💬 0 📌 0
Thank you so much for putting this together! This will help us design our next experiments. I'd be very curious to see how GEM-X compares to Flex head-to-head. I foresee that Flex will better detect lowly expressed genes (such as TF),...
18.03.2025 22:43 — 👍 3 🔁 0 💬 1 📌 0
I wanted to write briefly about a very pleasant experience we recently had coordinating and collaborating closely on competing publications with 2 other teams. 1/
24.01.2025 19:36 — 👍 114 🔁 23 💬 2 📌 5
@anusri.bsky.social first author & developer of ChromBPNet is looking for opportunities in industry in ML for bio/genomics. She is an excellent rigorous scientist (as u can see from the paper). Very strongly recommend her. Plz reach out to her if u have openings. Plz forward.
13.01.2025 18:23 — 👍 59 🔁 19 💬 3 📌 1

Another day, another #HRHorrorStory
08.01.2025 18:01 — 👍 0 🔁 0 💬 0 📌 0
Cant wait to read it😍
Felicitats Pau!!
04.01.2025 00:51 — 👍 1 🔁 0 💬 1 📌 0

One thing I didn't expect in the US is that I'd pay almost a third of my salary in taxes :(
This morning, the FICA taxes kicked in..
03.01.2025 18:55 — 👍 0 🔁 0 💬 0 📌 0
Our ChromBPNet preprint out!
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
25.12.2024 23:48 — 👍 231 🔁 89 💬 8 📌 5
Totally agree, but in practical terms that's what's gonna drive change
23.12.2024 04:41 — 👍 0 🔁 0 💬 1 📌 0
Of course, the main push back against this is: "if I publish my data I'll get scooped". And I totally sympathize with that, especially when people's careers are on the line. So we need to think as a community of ways of tackling this
23.12.2024 04:33 — 👍 0 🔁 0 💬 1 📌 0
Agreed! Not only that, pushing yourself to share code and data with the preprint ensures that it's properly done and not as an after thought or because "reviewers forced me too". What the latter accomplishes is GitHub repos that are impossible to navigate and data that is improperly annotated
23.12.2024 04:33 — 👍 1 🔁 0 💬 1 📌 0
Sharing code and data should also be required in the preprint. People are starting to use preprints as a way of advertising their science before it's published, sometimes with half-baked results. We should subject preprints to a higher level of rigor, with fully reusable, transparent and open data.
23.12.2024 04:25 — 👍 2 🔁 0 💬 1 📌 0
Some of the best ways to take advantage of preprints: share broadly that you posted one but also to send your preprint directly to colleagues who might care and may have missed it. Just as you like to be alerted to research you care about and may not have seen yet, so does everyone else
22.12.2024 22:21 — 👍 17 🔁 2 💬 1 📌 1
Still in 2024, I see way too many papers that don't share their data openly. I understand that in some cases the raw genomic reads need to have restricted access, but there's no excuse not to share any count matrix or processed object openly in repositories such as FigShare or Zenodo.
18.12.2024 17:00 — 👍 1 🔁 0 💬 0 📌 0
Felicitats Pau!! Molt bona feina!!
25.11.2024 22:25 — 👍 1 🔁 0 💬 0 📌 0
Hello, world!
25.11.2024 02:28 — 👍 4 🔁 0 💬 1 📌 0
Professor at #EPFL - #RNAVelocity inventor - Laboratory of Brain Development and Biological Data Science. Single-cell and spatial biologist studying brain development and lipids. #ERCStg investigator
🇨🇱👩🔬 PhD in Medical Sciences - Skin immunology | Postdoc in T cells & Cancer immunology at Weill Cornell Medicine 🗽| Pew Latin American Fellow 2024 | Biochemist | 🌍🚀🏐🐶🐱👩🍳🍀🌵
Molecular cell biology and stats / ML, mostly scRNA-seq.
Principal Scientist at Tahoe Tx.
https://www.nxn.se/archive
Mentor, scientist & engineer. Having fun in @slavovlab.bsky.social and Parallel Squared Technology Institute @parallelsq.bsky.social with biology & single-cell proteomics.
https://nikolai.slavovlab.net
Bioinformatics Scientist | Multi-omics | Cancer
Pathology resident & postdoc @jhu.edu studying pancreatic & colorectal precancers | Emory & Tufts alum | Dad, husband, Mainer, Boston sports fan
Dad and husband spending most of my time thinking about cancer. Math modeler. Convergence Institute at Johns Hopkins. Postdoc.
Instructor at DFCI | Multiple Myeloma | T-cell Therapies | Spatial Biology
Scientist working to defeat pancreatic cancer. Co-director of the Brenden-Colson Center for Pancreatic Care. Oregon Health and Science University
Director of Cancer Biology at Sage Bionetworks. Building radically collaborative cancer research communities through team science and the integration of clinical, multiomic, single-cell, & spatial data with the HTAN DCC, MC2 Center, & AACR Project GENIE.
Postdoctoral Scholar at University of California, San Francisco | Skin Biology | Skin Cell and Cancer Genomics and Transcriptomics | Stem Cell Biology | Painting | Hiking
Associate Professor @mdanderson.bsky.social | Single-Cell Spatial Omics, Computational Biology, Genomics, Immunology, Data Science | Advancing Precision & Predictive Oncology
Scientist, believer in truth and justice, lover of books and science journals
Pediatric oncologist and physician-scientist | Opinions my own
We use tumor genomics 🧬 and #zebrafish models 🎏 to develop new approaches for childhood cancers #sarcoma #Wilmstumor #GCT
Assistant Professor @harvard • Tumor Microenvironment | Spatial Biology | Computational | Multimodal Data Integration • PI at @nirmallab.com
Senior Investigator @ Altius Institute for Biomedical Sciences. Research: High-resolution mapping of chromatin structure & function. Fun: Mountain shenanigans and skiing turns all year. Seattle, USA/Patagonia Chilena (🇺🇸🇨🇱). http://vierstra.org