
The Open Molecules 2025 (OMol25) Dataset, Evaluations, and Models
Machine learning (ML) models hold the promise of transforming atomic simulations by delivering quantum chemical accuracy at a fraction of the computational cost. Realization of this potential would en...
It's finally done (enough for a preprint)! Today, in collaboration with so many folks at Meta (shout-out Daniel Levine and Muhammed Shuaibi, who put in superhuman levels of work), Berkeley, Stanford, NYU, and more, I'm proud to announce the Open Molecules 2025 (OMol25) dataset!
#CompChem #ML 🧪 ⚗️
14.05.2025 16:06 — 👍 39 🔁 10 💬 1 📌 0

The phrase "pathologically disorganized" is used to describe a neural network trained to predict protein structure from sequence embeddings
A post by @ncfrey.bsky.social and @amyxlu.bsky.social on repurposing ESMFold for protein design, featuring one of my favorite phrases in the field ncfrey.substack.com/p/hit-the-vi...
19.03.2025 16:51 — 👍 15 🔁 4 💬 0 📌 0
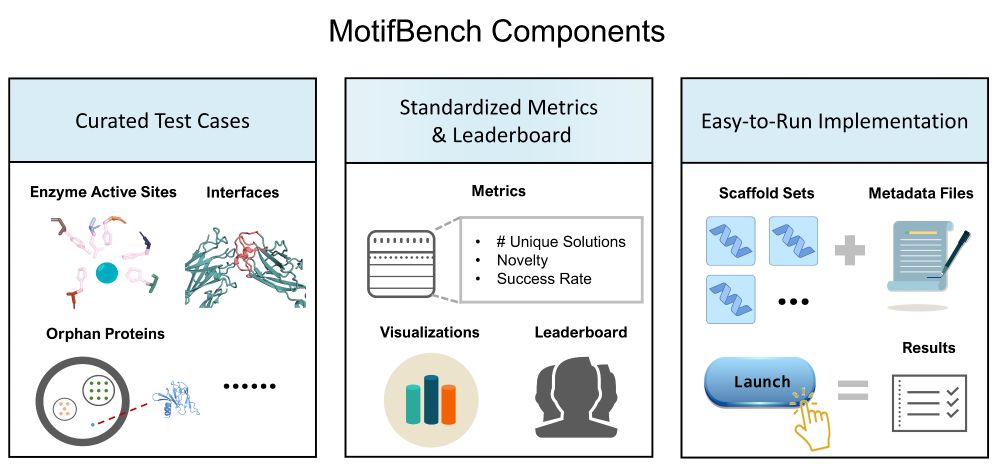
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
19.02.2025 20:49 — 👍 41 🔁 17 💬 1 📌 5
This was a massive, 3+ effort with 60+ collaborators, and we're still only scratching the surface of what's possible in biology, drug design, and ML. Incredibly proud of this team and honored to be a part of this research and sharing it with the world.
26.02.2025 21:15 — 👍 1 🔁 0 💬 0 📌 0
We @prescientdesign.bsky.social Genentech pre-printed our "Lab-in-the-loop for therapeutic antibody design." We built a general ML system to accelerate molecule design for challenging, therapeutically relevant targets.
www.biorxiv.org/content/10.1...
26.02.2025 21:15 — 👍 23 🔁 8 💬 1 📌 2

Language Models Optimize Biologically Realistic Synthetic Sequences, Potentially Helping Drug…
CDS researchers unveil LLOME, which optimizes biologically realistic synthetic sequences with potential applications for drug discovery.
CDS PhD student @angie-chen.bsky.social presents LLOME, using LLMs to optimize synthetic sequences with potential applications for drug design.
Co-led by @activelearner.bsky.social & @ncfrey.bsky.social and others at @prescientdesign.bsky.social
nyudatascience.medium.com/language-mod...
15.01.2025 19:44 — 👍 9 🔁 3 💬 0 📌 0

New blog post with Aya Ismail on our recent work, which introduces a fundamentally new way to build foundation models that are interpretable by design for scientific discovery.
tinyurl.com/cb-plm-blog
12.12.2024 18:36 — 👍 8 🔁 3 💬 1 📌 0
My team @prescientdesign.bsky.social is hiring! 🎉
Join me, @stephenra.com, @keunwoochoi.bsky.social, @kyunghyuncho.bsky.social, and the Large Molecule Drug Discovery AI/ML and LLM teams to work on basic research and applications of LLMs to drug discovery.
Link to apply: tinyurl.com/prescient-lmdd
11.12.2024 18:25 — 👍 6 🔁 0 💬 0 📌 0
Incredible work led by @amyxlu.bsky.social introducing PLAID, an all-atom co-generation method for proteins that requires only sequence inputs for training data! Read Amy's thread below, with links to the preprint, code, and model weights!
👇
06.12.2024 19:26 — 👍 6 🔁 0 💬 0 📌 0
get in early on the best podcast for bio, chem, and ML. fill the void in your life of entertaining, technical content made by experts for experts (and enthusiasts!).
04.12.2024 23:02 — 👍 6 🔁 0 💬 0 📌 0

A common question nowadays: Which is better, diffusion or flow matching? 🤔
Our answer: They’re two sides of the same coin. We wrote a blog post to show how diffusion models and Gaussian flow matching are equivalent. That’s great: It means you can use them interchangeably.
02.12.2024 18:45 — 👍 254 🔁 58 💬 6 📌 7

Overview of the BioM3 framework for protein design via natural language prompts.

Wet-lab validation
BioM3: a model that generates proteins conditioned on text prompts, with wet-lab validation!
www.biorxiv.org/content/10.1...
25.11.2024 22:38 — 👍 80 🔁 15 💬 1 📌 1
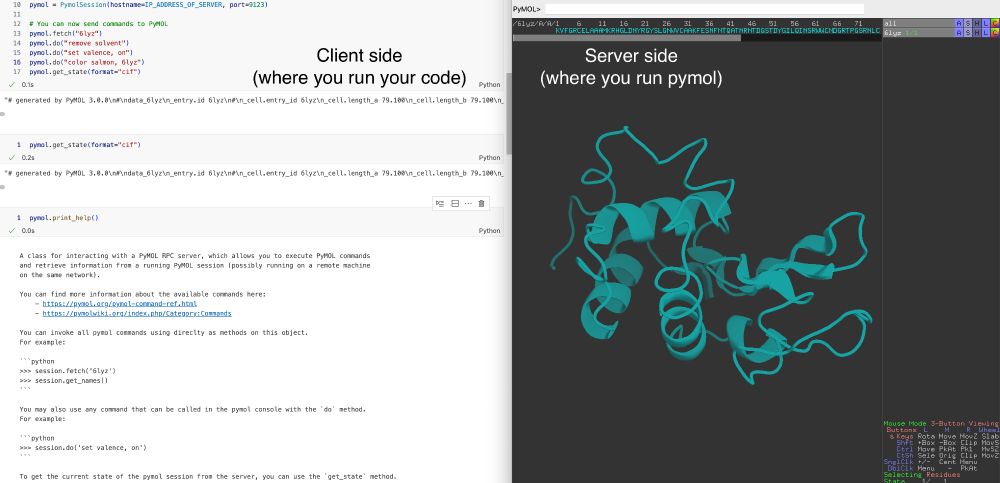
A weekend project from a while back -- this little package (with no dependencies) allows you to interact with pymol remotely.
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
25.11.2024 14:50 — 👍 47 🔁 13 💬 4 📌 3
The first list filled up, so here's a second list of AI for Science researchers on bluesky.
Let me know if I missed you / if you'd like to join!
bsky.app/starter-pack...
20.11.2024 08:56 — 👍 70 🔁 29 💬 58 📌 0
I'm making a list of AI for Science researchers on bluesky — let me know if I missed you / if you'd like to join!
go.bsky.app/AcP9Lix
10.11.2024 00:11 — 👍 246 🔁 90 💬 160 📌 5
🧪 For people interested in AI & enzymes (enzyme engineering, design, discovery, ...), I'm assembling a starter pack for us.
DM if you'd like to be included!
go.bsky.app/MhfaQBh
20.11.2024 10:29 — 👍 36 🔁 8 💬 6 📌 1
bsky.app/profile/kevi...
19.11.2024 01:34 — 👍 4 🔁 0 💬 0 📌 0
Two BioML starter packs now:
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
DM if you want to be included (or nominate people who should be!)
18.11.2024 17:09 — 👍 119 🔁 56 💬 10 📌 11
In a gratuitous attempt to acquire more followers myself 😁, I've made a start on a "starter pack". Hopefully as more people from 🐦 make it over to 🦋, we can extend this a bit. Suggestions welcome!
I've noticed not all accounts seem to be eligible to be added, anyone know what's up with that? 🤔
15.11.2024 20:04 — 👍 125 🔁 37 💬 34 📌 10
I tried to make a bioml starter pack. DM if you want me to add or remove you?
go.bsky.app/2VWBcCd
11.11.2024 23:45 — 👍 89 🔁 39 💬 29 📌 6
I’ve started an AI in healthcare starter pack! Let me know who is missing. go.bsky.app/7PeNwep
08.11.2024 05:01 — 👍 111 🔁 39 💬 53 📌 3
Made a biotech starter pack because I want to meme with y'all on this site instead of the old one
go.bsky.app/TbKxUEk 🧪🧬💻
09.11.2024 21:31 — 👍 106 🔁 25 💬 16 📌 6
👋
16.11.2024 14:26 — 👍 2 🔁 0 💬 0 📌 0
About Me
About Me
Hello BlueSky! 👋 I'm Nathan, a scientist at Prescient Design • Genentech. I lead an incredibly talented team of researchers working to transform drug discovery through computation, AI/ML, engineering, and data-centric thinking.
Find out more about our team and our work on ncfrey.github.io
16.11.2024 13:55 — 👍 19 🔁 2 💬 1 📌 0
Assistant Professor at Stanford Statistics and Stanford Data Science | Previously postdoc at UW Institute for Protein Design and Columbia. PhD from MIT.
House of Cinema podcast
Making videos and what not
Tron Legacy enthusiast
Research Scientist @prescientdesign @Genentech Former PhD @umdcs
ayaismail.com
A Genentech Accelerator
gene.com/prescient
https://scholar.google.com/citations?user=HVfvEVoAAAAJ
Purveyor of Dismissed Cinema | PanCan Survivor | Author | Fat Lesbian Wife | Fangoria & /Film | Co-Host of This Ends at Prom Podcast | Mouthy Midwest Broad in L.A.
Immigrant. Tech/movie 🇬🇾 guy. Engadget Editor. Filmcast co-host. ATL Film Critics Circle member. He/Him. devindra@devindra.org @devindra@mastodon.social
Movies: thefilmcast.com
Video Games: dlcpod.com
Science: wehaveconcerns.com
Books: youtube.com/cannatajeff
I love loving things.
I’ve been reviewing movies and TV on the internet for 15 years. I host podcasts like Decoding TV and The Filmcast. Check out my newsletter: www.Decodingeverything.com
Researcher at Google DeepMind in London. Previously PhD at Cambridge University.
Postdoc at MIT. Generative models, inference, AI for science. Prev: Princeton, Meta, NUS. liusulin.github.io
Professional Stumbler. Shenanigans with Earth Science & Machine Learning
Visiting researcher at google & assistant prof at it:u (at)
Lecturer (Asst. Prof.) in meteorology and math. & stats | Data assimilation, climate, ML, applied dynamical systems, predictability, inverse problems, climate politics
She/her 🏳️⚧️
https://eviatarbach.com
Book Author - AI for Science
Senior AI PM @HelloBetter
Open-ended scientific discovery
Animal rights activist
PhD research fellow at Active Inference Institute, data science, cognitive science, probabilistic models, Julia, Montana regenerative hobby farmer, author of Economic Direct Democracy: A Framework to End Poverty and Maximize Well-Being. #wellbeingeconomy
Thanks for coming to “Foundation Models for Biological Discoveries” (FMs4Bio) @ AAAI 2025!
Ph.D. Student @ ELLIS Unit / University Linz Institute for Machine Learning
Assistant Professor of Computer Science at the University of Virginia. I work on Responsible AI (differential privacy & fairness) and machine learning for science and engineering (differentiable optimization) | http://nandofioretto.github.io
Co-Founder & Chief Scientist @ Emmi AI. Ass. Prof / Group Lead @jkulinz. Former MSFTResearch, UvA_Amsterdam, CERN, TU_Wien
AI faculty @ ELLIS Institute Tübingen & Max Planck Institute for Intelligent Systems. Heading the Computational Applied Mathematics & AI Lab (CAMAIL)
Prev: MIT CSAIL + ETH Zurich
https://camail.org/
Machine learning for molecular biology. ELLIS PhD student at Fabian Theis lab. EPFL alumnus.










