We are super thankful to everyone who contributed to this massive team effort, and especially Jeff Hawkes, Carsten Simon, Bruno Brandao da Costa, and Jarmo-Charles Kalinski for coordinating and spearheading the study and paper.
07.02.2026 18:33 —
👍 0
🔁 0
💬 0
📌 0
The very positive aspect from my perspective: This work highlights the power of open science and large-scale collaboration to move non-targeted metabolomics forward.
07.02.2026 18:32 —
👍 0
🔁 0
💬 0
📌 0
There is still room for improvement on data alignment side, and we as community should put more emphasis on method standardization, especially if long-term data comparability and reuse is desired.
07.02.2026 18:32 —
👍 1
🔁 0
💬 0
📌 0
Remaining challenges are: That low-intensity signals in DOM showed higher variability, especially for DDA-based MS/MS acquisition. Not so surprisingly, differences in instrument acquisition rates and standardization strongly influence comparability.
07.02.2026 18:31 —
👍 0
🔁 0
💬 0
📌 0
The good news is: Data from similar MS platforms with harmonized parameters reveal consistent chemical trends, and high-intensity features, and multivariate analysis aligned well across labs.
07.02.2026 18:30 —
👍 0
🔁 0
💬 0
📌 0
This project brought together 50+ co-authors from 24 laboratories, all analyzing identical DOM samples using standardized LC and MS/MS settings. The goal was to check if we all find the same molecules and chemical trends, and to assess how well we can co-analyze data from different labs.
07.02.2026 18:30 —
👍 0
🔁 0
💬 0
📌 0

I am super excited that our paper on inter-laboratory comparability of non-targeted LC–MS/MS analysis of dissolved organic matter was just published in ES&T.
pubs.acs.org/doi/10.1021/...
07.02.2026 18:28 —
👍 15
🔁 5
💬 6
📌 0

Mapping of the viral shunt across widespread coccolithophore blooms using metabolic biomarkers | PNAS
The viral shunt is a fundamental ecosystem process which diverts the flux of organic
carbon fixed through photosynthesis during algal bloom events ...
🌊Paper announcement! 📣
Viral infections rewire the metabolic makeup of their host and thereby create distinct chemical signatures. Can we use metabolic biomarkers to diagnose infections of algal blooms in the ocean?
Well, take a look at our new article led by Conny Kuhlisch in @pnas.org
>>
05.02.2026 17:22 —
👍 28
🔁 15
💬 2
📌 1
let's get the community rolling 💪! Thanks to Vilhelm Suksi from @antagomir.bsky.social 's group for the contribution of the notame vignette 🙌
07.01.2026 15:25 —
👍 3
🔁 2
💬 0
📌 0

xcms in Peak Form: Now Anchoring a Complete Metabolomics Data Preprocessing and Analysis Software Ecosystem
High-quality data preprocessing is essential for untargeted metabolomics experiments, where increasing data set scale and complexity demand adaptable, robust, and reproducible software solutions. Modern preprocessing tools must evolve to integrate seamlessly with downstream analysis platforms, ensuring efficient and streamlined workflows. Since its introduction in 2005, the xcms R package has become one of the most widely used tools for LC-MS data preprocessing. Developed through an open-source, community-driven approach, xcms maintains long-term stability while continuously expanding its capabilities and accessibility. We present recent advancements that position xcms as a central component of a modular and interoperable software ecosystem for metabolomics data analysis. Key improvements include enhanced scalability, enabling the processing of large-scale experiments with thousands of samples on standard computing hardware. These developments empower users to build comprehensive, customizable, and reproducible workflows tailored to diverse experimental designs and analytical needs. An expanding collection of tutorials, documentation, and teaching materials further supports both new and experienced users in leveraging broader R and Bioconductor ecosystems. These resources facilitate the integration of statistical modeling, visualization tools, and domain-specific packages, extending the reach and impact of xcms workflows. Together, these enhancements solidify xcms as a cornerstone of modern metabolomics research.
Out now! xcms in Peak Form: Now Anchoring a Complete Metabolomics Data Preprocessing and Analysis Software Ecosystem doi.org/10.1021/acs....
with Phillipine and @jorainer.bsky.social (EURAC), @metabomichael.bsky.social, Hendrik and Norman from @ipbhalle.bsky.social, @janstanstrup.bsky.social, et al.
08.12.2025 20:26 —
👍 25
🔁 10
💬 1
📌 1

We are super excited that our new paper on microfluidic-based LC-MS/MS fractionation in combination with bioluminescence bioreporters readouts, for compound-resolved bioactivity metabolomics, was just published: pubs.acs.org/doi/10.1021/...
26.10.2025 21:24 —
👍 15
🔁 7
💬 7
📌 0
What a fun day! Thanks so much for stopping by Pieter!
27.09.2025 16:58 —
👍 6
🔁 0
💬 0
📌 0

In a large community effort with 50 coauthors, we analyzed the same set of marine dissolved organic matter samples across 24 laboratories via non-targeted LC-MS/MS, to check if we get comparable data.
If you want to check it out, you can find our preprint here:
doi.org/10.26434/che...
17.09.2025 15:20 —
👍 7
🔁 1
💬 0
📌 0
Thanks! And yes, depending on the reagent and pH, you will have some ion suppression and the sensitive will drop. To bypass that, I would run the initial runs without infusion/pH modulation.
29.07.2025 14:09 —
👍 0
🔁 0
💬 0
📌 0
Thanks so much to everybody who made this interdisciplinary project possible. And epically the editor and the three reviewers at @natcomms.nature.com for their throughout positive feedback.
27.07.2025 18:41 —
👍 5
🔁 3
💬 1
📌 0
Implementing the MCheM setup is pretty easy, and all reagents and hardware components are commercially available and relatively cheap.
MCheM data analysis is supported in @mzmine.bsky.social @gnps2.bsky.social and SIRIUS (@brightgiant.bsky.social) and free for academic researchers.
27.07.2025 18:36 —
👍 3
🔁 2
💬 0
📌 0
Thanks Gabriel :)
05.07.2025 16:17 —
👍 1
🔁 0
💬 0
📌 0
Thanks Don! Will post about the new work here and on our webpage www.functional-metabolomics.com/publication
04.07.2025 05:57 —
👍 1
🔁 0
💬 0
📌 0
Thanks a lot Tri :)
04.07.2025 05:52 —
👍 1
🔁 0
💬 0
📌 0
Thanks a lot Manuel!
04.07.2025 05:52 —
👍 0
🔁 0
💬 0
📌 0
Super excited that I’ve been selected as a Simons Early Career Investigator in Aquatic Microbial Ecology and Evolution. We will explore how marine microbes shape the production, transformation, and fate of dissolved organic matter.
Thanks so much @simonsfoundation.org
We can’t wait to get started!
03.07.2025 16:18 —
👍 28
🔁 4
💬 4
📌 0
Whoops, we couldn't find that.
www.grainger.com/product/SMC-...
I looked into it for a long time, happy to chat about details. I was close to buying one, but acilities finally increased the pressure to 110 psi. Was definitely worth the fight. Passive N2 generator works like a charm.
28.06.2025 06:12 —
👍 0
🔁 0
💬 1
📌 0
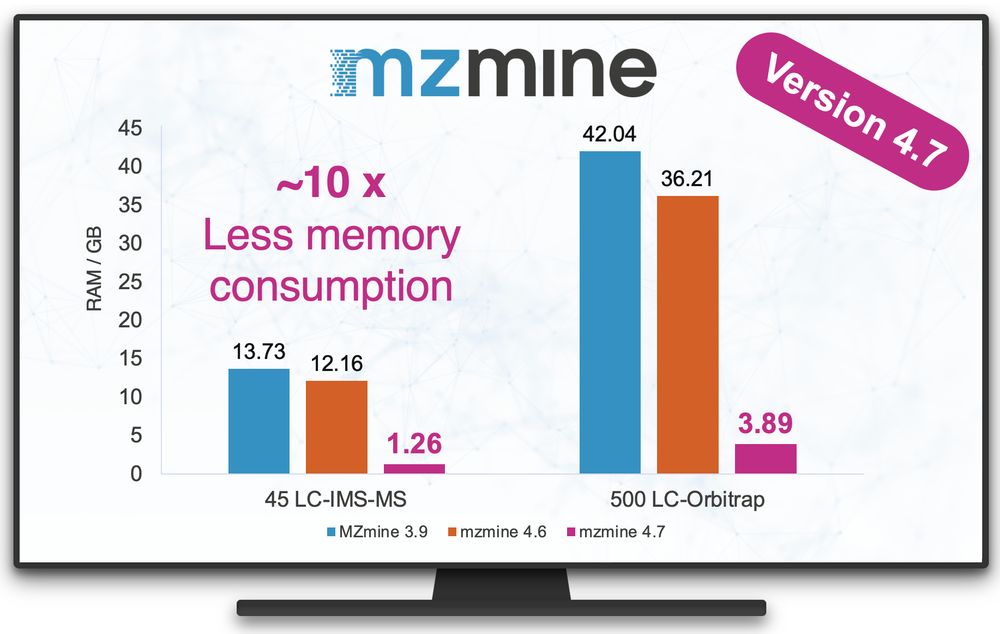
#mzmine 4.7 is now available!
This release brings our most significant improvement in memory efficiency to date, unlocking new capabilities for analyzing large-scale datasets.
Join us for a live software demo at our booth today and tomorrow at 12:00/noon during #ASMS2025
03.06.2025 14:20 —
👍 13
🔁 6
💬 0
📌 0
We are pretty stoked that our paper on chemical shifts in kelp forests in the Gulf of Maine made it onto the front cover of @science.org
Big congratulations to Shane Farrell and everybody involved! ✌️✌️✌️
22.05.2025 23:03 —
👍 12
🔁 2
💬 2
📌 0
CMFI Enters Second Funding Period | CMFI News
The members of CMFI can breathe a sigh of relief: On January 1, 2026, the Tübingen Cluster of Excellence will enter its second funding period with a duration of seven years. This was announced by the ...
🎉 Fantastic News 🎉
Our CMFI Cluster of Excellence @unituebingen.bsky.social receives funding extension for the next seven years.
Spokesperson Andreas Peschel @andreaspeschel.bsky.social: "We can now advance our research into resistance mechanisms and new antimicrobial agents!"
shorturl.at/rcK1A
22.05.2025 15:37 —
👍 17
🔁 3
💬 0
📌 3










