A multigenic quantitative trait locus underlies natural variation in Arabidopsis thaliana root system architecture and transcriptional responses to microbiota-derived Pseudomonas https://www.biorxiv.org/content/10.1101/2025.09.03.673899v1
05.09.2025 14:07 — 👍 1 🔁 2 💬 0 📌 0
This is sad news, but what a long and rich life Julian has had.
A towering and inspiring figure in microbiology and antimicrobial resistance.
06.02.2025 08:36 — 👍 15 🔁 5 💬 0 📌 1
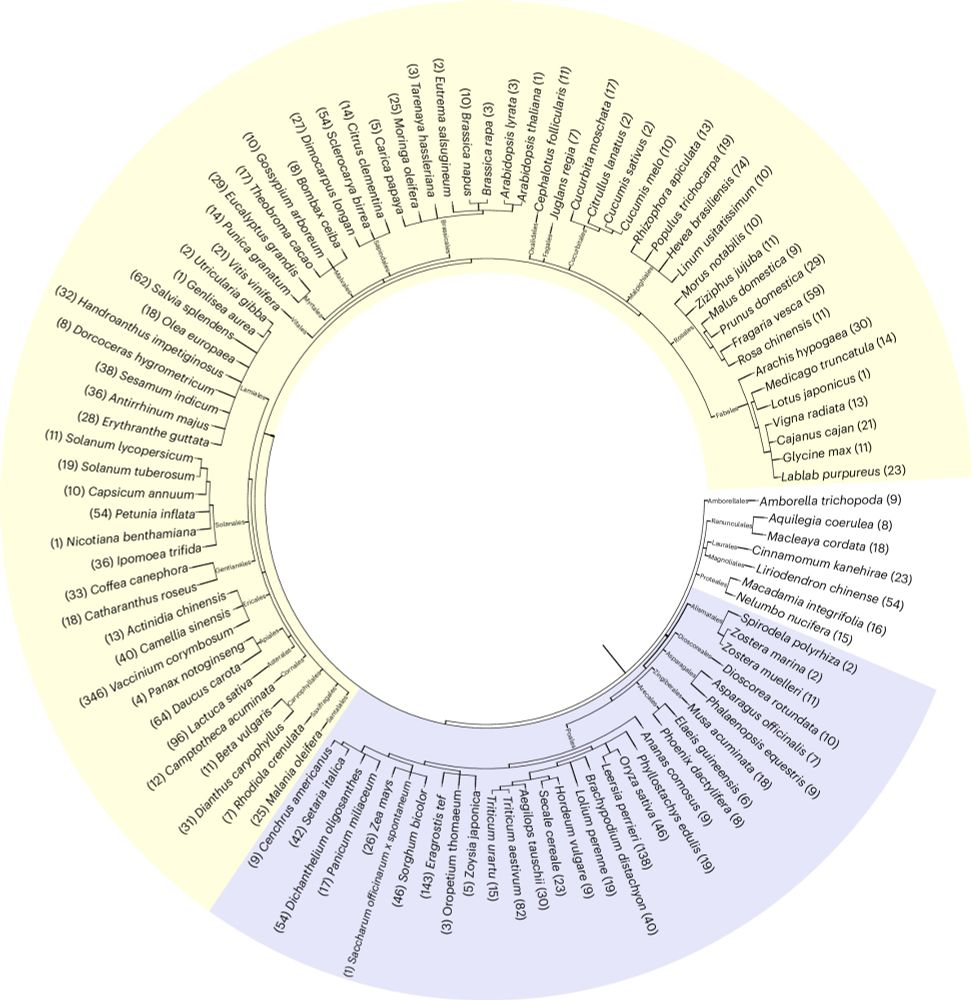
Tandem kinase proteins across the plant kingdom
Nature Genetics - This genomic analysis of tandem kinase proteins across 104 plant species highlights their mechanisms of convergent molecular evolution and potential roles in plant immunity.
First paper of the year is out! A collaboration with Tzion Fahima @University of Haifa with heavy lifting by Tamara Reveguk, who was a pleasure to co-supervise. We generated an atlas of tandem kinase proteins (TKPs). TKPs are known to confer disease resistance in monocots. 🧵(1/4)
rdcu.be/d5ADS
08.01.2025 22:08 — 👍 43 🔁 20 💬 4 📌 2

Starship giant transposons dominate plastic genomic regions in a fungal plant pathogen and drive virulence evolution
Starships form a recently discovered superfamily of giant transposons in Pezizomycotina fungi, implicated in mediating horizontal transfer of diverse cargo genes between fungal genomes. Their elusive nature has long obscured their significance, and their impact on genome evolution remains poorly understood. Here, we reveal a surprising abundance and diversity of Starships in the phytopathogenic fungus Verticillium dahliae. Remarkably, Starships dominate the plastic genomic compartments involved in host colonization, are enriched in virulence-associated genes, and exhibit genetic and epigenetic characteristics associated with adaptive genome evolution. We further uncover extensive horizontal transfer of Starships between Verticillium species and, strikingly, from distantly related Fusarium fungi. Finally, we demonstrate how Starship activity facilitated the de novo formation of a novel virulence gene. Our findings illuminate the profound influence of Starship dynamics on fungal genome evolution and the development of virulence. ### Competing Interest Statement The authors have declared no competing interest.
📣New year, new preprint @biorxiv-microbiol.bsky.social 🎉! A study led by the brilliant @yukiyosato.bsky.social showing that Starship giant transposons dominate plastic genomic regions in the fungal plant pathogen Verticillium dahliae and drive virulence evolution. 🧵[1/12] doi.org/10.1101/2025...
10.01.2025 14:23 — 👍 115 🔁 68 💬 3 📌 2
Quick plug for preview article Brian Kvitko and I wrote for Cell Host and Microbe (setting up an interesting paper from Grenz et al. in same issue) and waxing philosophical about subjectivity of host range in bacterial phytopathogens like Pseudomonas syringae
authors.elsevier.com/a/1kPEb6t8JE...
08.01.2025 16:21 — 👍 6 🔁 7 💬 0 📌 0

New Preprint! We identified Pseudomonas in wild liverworts and explored what makes them virulent. www.biorxiv.org/content/10.1...
03.12.2024 15:40 — 👍 103 🔁 49 💬 10 📌 6
📌
04.12.2024 02:31 — 👍 0 🔁 0 💬 0 📌 0
📌
20.11.2024 00:31 — 👍 0 🔁 0 💬 0 📌 0
📌
17.11.2024 16:19 — 👍 0 🔁 0 💬 0 📌 0
We are an independent, international centre of excellence in plant science, genetics and microbiology.
www.jic.ac.uk
Associate professor of Bioinformatics at Chongqing Medical University, China. Lab: https://mbio.info Personal: http://shenwei.me/ http://shenwei356.bsky.social
Writing a book about horizontal gene transfer and non treelike evolution. Bioinformatics, Evolutionary Biology. Pangenomes. Chair in Evolutionary Biology. 🇮🇪 http://github.com/mol-evol/panGPT
BlueSky account for https://botany.one the weblog of the Annals of Botany Company. Non-Profit All Science since 1887.
Posts by @alunsalt.bsky.social or @caordonezparra.bsky.social and scheduled well in advance through Buffer.
Advancing research and teaching in the most dynamic and vital areas of biological research
Assistant Professor UofT Cell & Systems Biology - 🐟🐠🐡 comparative genomics/ethology & evolutionary cell biology of sleep 💤 😴 + adventures with @hilsawh - https://csb.utoronto.ca/faculty/maxwell-shafer/
At our institute we study fundamental processes of plant physiology and development and the interaction of plants with their environment.
Post-doctoral researcher in the group of Prof. Marcel van der Heijden (University of Zürich, Switzerland).
Fungal biology | Soil & plant microbiota | Ecology & evolution | Plant-microbe-microbe interactions | Genomics | ...
Presidential Associate Professor @upenn.bsky.social - using AI to reimagine antibiotic discovery and peptide design. Previously @MIT, @UBC
🔗 https://delafuentelab.seas.upenn.edu
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
Postdoc in Heitman Lab @dukemedschool.bsky.social.
🌈Biology | 🍄🧬Fungal Genetics | 🦠#IDSky
👶Dad | 🎥Love movies | 🧱LEGO collector.
Why waste time say lot word when few word do trick?
Department of Natural product biosynthesis at the Max Planck Institute for Chemical Ecology in Jena, Germany
https://www.sarahoconnor.org/
HHMI Hanna Gray Fellow | Asst. Professor @ WashU & Asst. Member @ Danforth Center | Spatial Biology | Plant-microbe Interactions | Duckweed | YouTuber/Streamer | www.thecoxlab.org/
Systems Microbial Ecologist | Design Thinker | F1 & IndyCar fan. Metalhead. Studying microbes in Desert Soil and nearshore Marine Sediments.
Blog: uncultured.carinilab.com
Web: CariniLab.com
Linkedin: https://www.linkedin.com/in/paul-carini/
Ecological modelling; ecosystem resilience and stability to global change; international rep @ BESMacroecology SIG; AE @ Ecological Solutions & Evidence; Research Scientist @ Canadian Forest Service
Views and opinions are my own
Forest Fire Research Scientist with Canadian Forest Service 🔥🌲🌳 and the WildFireSat mission 🚀🛰 📡 | Advancing how we use satellites to monitor fires and their impacts | Woman in Remote Sensing | Mother in Science 💻👩💻👩🔬🧪 | she/hers | celiac | 📚🎵🎶👟☕️ | 🇨🇦🇺🇸
Husband, Dad, Entomologist, Ecologist, Researcher with NRCan Canadian Forest Service , and a (lapsed) Curler. Occasional climate scientist.
Forest Ecologist, Research Scientist at the Canadian Forest Service, musician, father, and co-Editor-in-Chief of the International Journal of Wildland Fire



