
Causal modelling of gene effects from regulators to programs to traits - Nature
Approaches combining genetic association and Perturb-seq data that link genetic variants to functional programs to traits are described.
GWAS has been an incredible discovery tool for human genetics: it regularly identifies *causal* links from 1000s of SNPs to any given trait. But mechanistic interpretation is usually difficult.
Our latest work on causal models for this is out yesterday:
www.nature.com/articles/s41...
A short🧵:
11.12.2025 17:54 — 👍 148 🔁 67 💬 3 📌 1

Programmatic design and editing of cis-regulatory elements
The development of modern genome editing and DNA synthesis has enabled researchers to edit DNA sequences with high precision but has left unsolved the problem of designing these edits. We introduce Le...
After a huge amount of work w/ @alex-stark.bsky.social's group, a new version of our Ledidi preprint is now out!
In an era of AI-designed proteins, the next leap will be controlling when, where, and how much of these proteins are expressed in living cells.
www.biorxiv.org/content/10.1...
10.12.2025 15:18 — 👍 48 🔁 21 💬 2 📌 0
Thank you Alex! Excited to see our paper published in @nature.com ! Huge thanks to @jeffspence.github.io , @tkyzeng.bsky.social , @emmamarydann.bsky.social, @nikhilmilind.dev, @marsonlab.bsky.social, @jkpritch.bsky.social, and all the members of the Pritchard and Marson labs for your enormous help!
11.12.2025 03:04 — 👍 27 🔁 13 💬 0 📌 0
New preprint from our lab, spearheaded by @maelledaunesse.bsky.social and in collaboration with Diego Villar!
In which we ask, can we detect joint signatures suggestive of positive selection on transcriptomes and epigenomes?
04.12.2025 16:50 — 👍 27 🔁 11 💬 2 📌 0
It was a pleasure meeting you. I really appreciated hearing about your work and enjoyed our conversation. Thank you for coming to Japan!
03.12.2025 14:26 — 👍 1 🔁 0 💬 0 📌 0

Happy to present TOGA2, developed by Yury Malovichko @ymalovichko.bsky.social, the faster, memory-efficient & more accurate TOGA1 successor (github.com/hillerlab/TO...). And annotations, orthologs & gene loss/dup data generated with 4 references for 883 placental mammal and with 5 refs for 676 ...
23.11.2025 21:49 — 👍 26 🔁 14 💬 1 📌 0

NEW pub in @science.org 🥳
Is it sponges (panels A & B) or comb jellies (C & D) that root the animal tree of life?
For over 15 years, #phylogenomic studies have been divided.
We provide new evidence suggesting that...
🔗: www.science.org/doi/10.1126/...
13.11.2025 20:33 — 👍 283 🔁 131 💬 14 📌 31

Excited to share Nona: a unifying multimodal masking framework for functional genomics.
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
10.11.2025 21:01 — 👍 33 🔁 13 💬 1 📌 2

Are you using any of our factor models, such as MOFA? 🛵
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
07.11.2025 10:29 — 👍 34 🔁 12 💬 1 📌 3

Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
05.11.2025 17:17 — 👍 205 🔁 77 💬 6 📌 11

New preprint led by the brilliant @aleksandra-marconi.bsky.social on cichlid brain diversification, fgf8a signalling and regulatory divergence with TEs on the mix! All part of a wonderful collaboration with @ebablab.bsky.social
www.biorxiv.org/content/10.1...
@camzoology.bsky.social
24.10.2025 20:45 — 👍 74 🔁 23 💬 5 📌 1
🧠🌟🐭 Excited to share some of my postdoc work on the evolution of dexterity!
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
22.10.2025 20:41 — 👍 370 🔁 123 💬 18 📌 26
Thank you!
17.10.2025 09:01 — 👍 1 🔁 0 💬 0 📌 0
Thank you so much, Mari 😊 I’m also really looking forward to seeing all the amazing science coming from your lab!
17.10.2025 07:51 — 👍 0 🔁 0 💬 0 📌 0
So excited to see this out on bioRxiv! Huge thanks to @kaessmannlab.bsky.social and the amazing cerebellum team (@marisepp.bsky.social and @ioansarr.bsky.social) for all the support and insightful discussions that made this work possible!!
17.10.2025 05:58 — 👍 14 🔁 1 💬 1 📌 1
Excited to share the preprint from my main postdoc project! It’s been a long journey—huge thanks to everyone who made it possible, especially @leticiarm1618.bsky.social for being the best collaborator one could ask for, and the amazing @kaessmannlab.bsky.social lab for the invaluable support!
15.10.2025 13:39 — 👍 13 🔁 6 💬 1 📌 0

🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
16.10.2025 06:23 — 👍 282 🔁 110 💬 7 📌 11
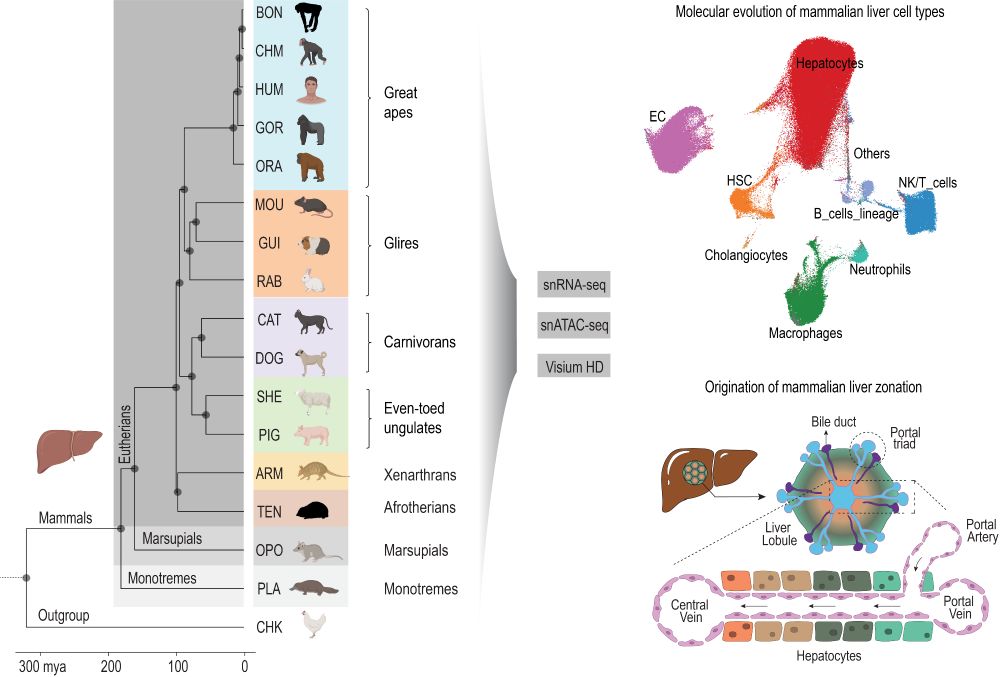
We are thrilled to share our new preprint entitled “The origin and molecular evolution of the mammalian liver cell architecture” www.biorxiv.org/content/10.1...
15.10.2025 12:51 — 👍 53 🔁 19 💬 3 📌 2

Which mutations rewire function of regulatory DNA?
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
09.10.2025 12:02 — 👍 29 🔁 10 💬 1 📌 0
New preprint from the lab! We discovered a transient fluidization in the basal region of human forebrains by tracking microdroplets in cerebral organoids.This “basal fluidization”, absent in gorilla and mouse, may contribute to greater surface expansion in human forebrains
1/
doi.org/10.1101/2025...
08.10.2025 13:05 — 👍 86 🔁 35 💬 3 📌 3
Happy to share work spearheaded by former grad student Colin Shew testing shared duplicated cis regulatory elements (CREs) using an MPRA. While we find some high effect CREs, collectively paralog differences represent modest effects accounting for observed gene expression divergence.
07.10.2025 02:31 — 👍 30 🔁 13 💬 3 📌 0

A human-specific regulatory mechanism revealed in a pre-implantation model
Nature - Genetic manipulation of blastoids reveals the role of recently emerged transposable elements and genes in human development.
Today in @nature.com, we present our work leveraging functional genomics and human blastoids to uncover a human-specific mechanism in preimplantation development driven by the endogenous retrovirus HERVK.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
01.10.2025 18:08 — 👍 135 🔁 50 💬 11 📌 5

New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
26.09.2025 18:06 — 👍 54 🔁 20 💬 3 📌 2

We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
22.09.2025 05:29 — 👍 174 🔁 90 💬 4 📌 5

bioRxiv - An unbiased survey of distal element-gene regulatory interactions with direct-capture targeted Perturb-seq
New preprint from our lab!
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/
19.09.2025 03:03 — 👍 57 🔁 17 💬 1 📌 1

Cell-extrinsic controls over neocortical neuron fate and diversity
Cell-extrinsic cues are key for neocortical cell identity and diversity.
Ever wondered how robust cellular identity is to external perturbations? Here we disrupt cellular environmnent in vivo and in vitro, and find cell population specific sensitivities. Environment sculpts development yes, but not all cells are made of the same wood. www.science.org/doi/10.1126/...
19.09.2025 06:04 — 👍 66 🔁 20 💬 6 📌 0
🧠🦈Excited to present our latest work🧠🦈Interested in brain evolution? And shark embryos? Then read on… Our work sheds light on the deep origins of our brain’s most complex regions.
02.09.2025 17:45 — 👍 97 🔁 29 💬 6 📌 3
🧬 Join our world-class courses, conferences, and workshops at the forefront of molecular life science and its applications. 👩🏻🔬 www.embl.org/events/ 🔬
PI @Rockefeller University. Investigator @HHMI. Instigator of clonal raider ant project #CRAP. 🐜 Evolution, Behavior & Neuroscience. Posts science and photography. 🧠 📸
https://www.rockefeller.edu/research/2280-kronauer-laboratory/
Academic Track Research Fellow at Newcastle University. Studying object manipulation by animals without hands 🐝🐥🪺 animal behaviour, ecology, cognition, evolution. Dog and food lover. Mum. Views my own. she/her https://shokosugasawa.weebly.com/
Head of the Comparative Functional Genomics lab @Pasteur.fr | @inserm.fr | @erc.europa.eu
Comparative omics - Reproductive evolution - Data witchcraft.
Posts mine alone, in English (mostly) & French. #Evolution #Bioinformatics #openaccess #EDI. Director of @dee_unil and group leader at @SIB. He/Him. PI of @bgeedb, of […]
🌉 bridged from ⁂ https://ecoevo.social/@marcrr, follow @ap.brid.gy to interact
Assistant Professor UTSW; Mutagenesis, Population genetics, Evolution in somatic tissues and on phylogenetic tree
Thinking about work, life, and balance. Looking for answers in the cerebellum.
Assistant Professor @ Virginia Tech
www.vanderheijdenlab.com
Neuroscientist. Professor at Harvard University.
Studies the neural mechanisms underlying decision-making and learning. Dopamine.
Pangenomics for studying genome variation and evolution | Postdoc at Langmead Lab, Johns Hopkins | Formerly at UNIL/SIB/WUR | Incoming Assistant Professor | #hiring PhD student |sinamajidian.github.io
Associate Professor at Erasmus MC. MD, PhD, Clinical Geneticist, interested in gene regulation and the non-coding genome, bridging research and patient care
VIB Center for AI & Computational Biology
🧑💻 Director @steinaerts.bsky.social
📣 We're recruiting group leaders vib.ai
📻 Follow us for all the latest updates
Chief Scientist RIKEN PRI, Assoc Prof Univ Tokyo Department of Physics, Institute for Physics of Intelligence. Biophysics, noneq statphys. https://noneq-biophys.riken.jp/
Group Leader @mrc_hgu investigating gene regulation in development & human disease
Genomics, AI, sequence-to-function models, mechanisms of the cis-regulatory code. Investigator at the Stowers Institute.
Evolutionary epigenomics ( eukaryotes / Transcription Factors / Transposable Elements / DNA methylation ) @ QMUL (London).
Lab website: https://www.demendozalab.com/
Fights like a cow.
taipalelab.org
postdoc @trono-lab.bsky.social at EPFL🇨🇭// Future PI at @igbmc.bsky.social 🇫🇷 // Fascinated in transposons 🤘 and embryogenesis 👶
Future Lab: https://orspf.github.io/Rosspopoff.Lab.io/
feeds: biorxiv, medrxiv, arxiv.q-bio, arxiv.cs, arxiv.math, Research Square
regex: '(single(- )(cell|nuclei)|s[cn]RNA-seq)'
If you have suggestions, please get in contact with @willmacnair :)
UC-Berkeley Postdoc🐻, Edison Scientific Consultant👨🏽💻, Evomics Workshop Codirector🧬
prev Vanderbilt PhD, FutureHouse, Latch, MantleBio (acquired)
🌲 https://linktr.ee/jlsteenwyk
📍 https://jlsteenwyk.com/
Researcher PI @CIRI_Lyon @CNRS @ENSdeLyon
Functional evolution of virus-host interactions. Impact on cross-species transmissions. 🦇 🦍 🦠 🧬 🌳




















