
🚀 Thinking about life beyond academia?
Join BrisJAMS and Australian Society of Microbiology Qld Branch, this November for an inspiring evening of stories, insights, and real-world advice from scientists who’ve successfully built careers outside the academic path!🌟
6 November 2025, 6:00 PM – 8:00 PM
27.10.2025 00:28 — 👍 3 🔁 3 💬 3 📌 1
Thanks Steve! It was great working with you on the BrisJAMS committee 🙂
15.09.2025 10:11 — 👍 0 🔁 0 💬 0 📌 0

We are starting a new series where we introduce our BrisJAMS committee memebers in a fun-microbiology-kinda way 😊 🔬 🦠 🧫 🧪
Starting with none other than our wonderful Director
- Meghann Thai 😎 😁
08.09.2025 01:48 — 👍 4 🔁 1 💬 1 📌 0

Looking for a postdoc that combines wet and dry lab experience?
The EMBL-EBI-Sanger postdoctoral fellowships might be the thing for you!
Choose one of our pre-defined topics or propose your own.
www.ebi.ac.uk/research/pos...
@sangerinstitute.bsky.social
#postdoc #sciencecareers
🧬🖥️🔬
04.09.2025 16:24 — 👍 21 🔁 15 💬 0 📌 0

We're looking for an awesome computational #postdoc to join our group @PioneerCampus. You'll explore native cells with #cryoET, get long-term funding (5+ years), and live in Munich! For more info, see our new Cell Architecture Lab website:...
13.08.2019 16:07 — 👍 11 🔁 10 💬 2 📌 1
📣 New paper alert! Just out in Cell Reports! pubmed.ncbi.nlm.nih.gov/40644298/
Thrilled to share that we have discovered a brand-new anti-phage defense system! Bacteria have evolved various defense strategies (CRISPR etc) to counter phage attacks. We found a new one - fascinating and dramatic
⚔️🦠❄️🔬
02.08.2025 15:01 — 👍 150 🔁 52 💬 3 📌 5
We (with Clement Coclet, not on Bsky) had the chance to work on a broad "state of viromics" review. We tried to use this to give an overview of how the field changed over the last ~ 15 years, and also what we think are some of the major remaining challenges. Full-text access at -> rdcu.be/excHt
22.07.2025 15:06 — 👍 84 🔁 41 💬 2 📌 2
Thank you Courtney! GenomeFISH should work on roots as well, as long as you can get the genomes of the microbes of interest for probe generation :)
17.07.2025 23:10 — 👍 1 🔁 0 💬 0 📌 0

Logo for the Sandpiper website
Out in @natbiotech.nature.com: Metagenome taxonomy profilers usually ignore unknown species. SingleM is an accurate profiler which doesn't, even detecting phyla with no MAGs. Profiles of 700,000 metagenomes at sandpiper.qut.edu.au. A 🧵
16.07.2025 21:59 — 👍 131 🔁 71 💬 7 📌 9
In the target population, the GenomeFISH and DAPI signal are positively correlated, resulting in a normalised signal intensity of ~1 when dividing the GenomeFISH signal by the DAPI signal. For closely related strains, this value drops below 1, as the GenomeFISH signal reduces in strength.
(2/2)
08.07.2025 22:58 — 👍 0 🔁 0 💬 0 📌 0
Thanks Nicholas! We normalise the GenomeFISH signal to the DAPI signal. As genome copy number and image plane orientation can all influence the signal strength in the target population and probes will also bind to closely related strains, normalisation is key.
(1/2)
08.07.2025 22:56 — 👍 0 🔁 0 💬 1 📌 0
Thanks Steve!
08.07.2025 21:01 — 👍 0 🔁 0 💬 0 📌 0
No need for design :) GenomeFISH probes are generated from single amplified genomes or isolate genomes by simply fragmenting the entire genome and fluorescently labelling the fragments.
08.07.2025 06:48 — 👍 1 🔁 0 💬 1 📌 0
Given the high-throughput nature of GenomeFISH, its superior sensitivity, and its phylogenetic resolution, we hope it will become widely used for the visualisation of complex microbial communities.
08.07.2025 02:33 — 👍 0 🔁 0 💬 0 📌 0
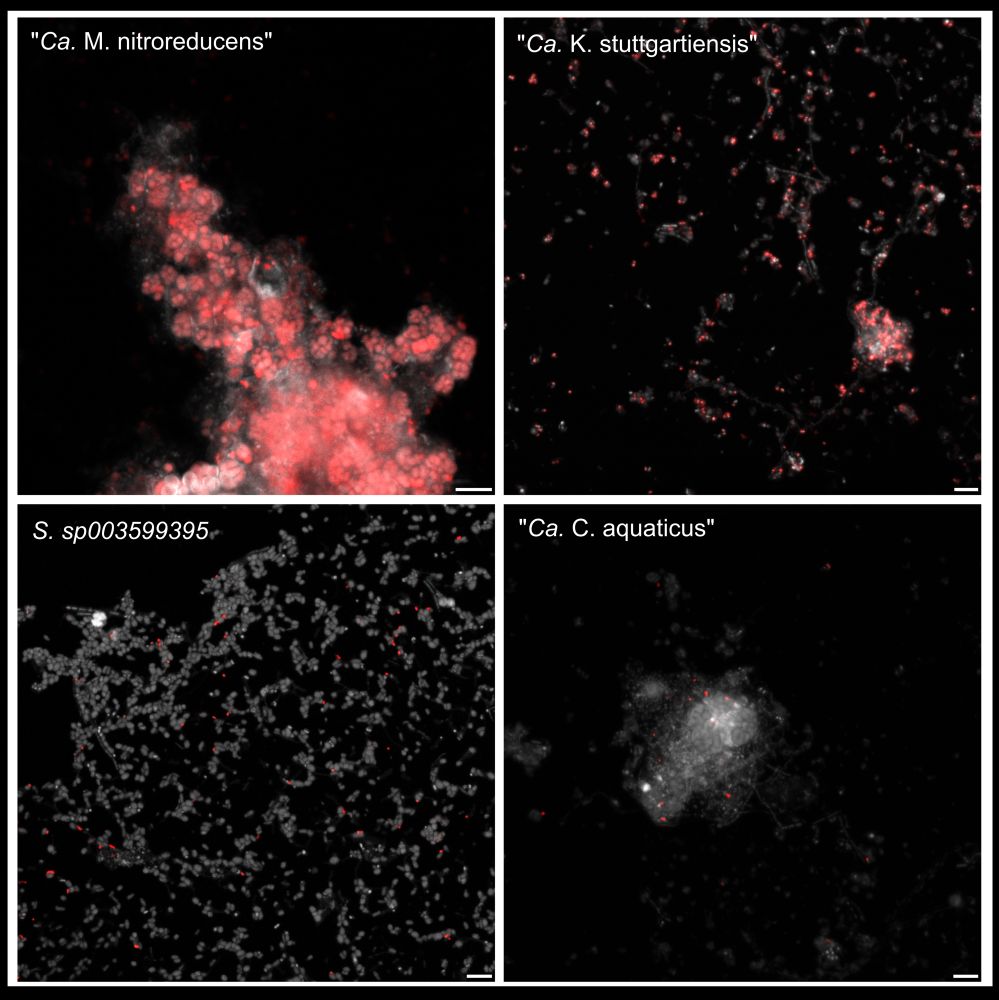
It also enables GenomeFISH to reach superior signal intensities compared to traditional FISH, allowing it to visualise microbial lineages that are challenging to visualise with traditional FISH, such as members of the Patescibacteria.
08.07.2025 02:32 — 👍 0 🔁 0 💬 1 📌 0
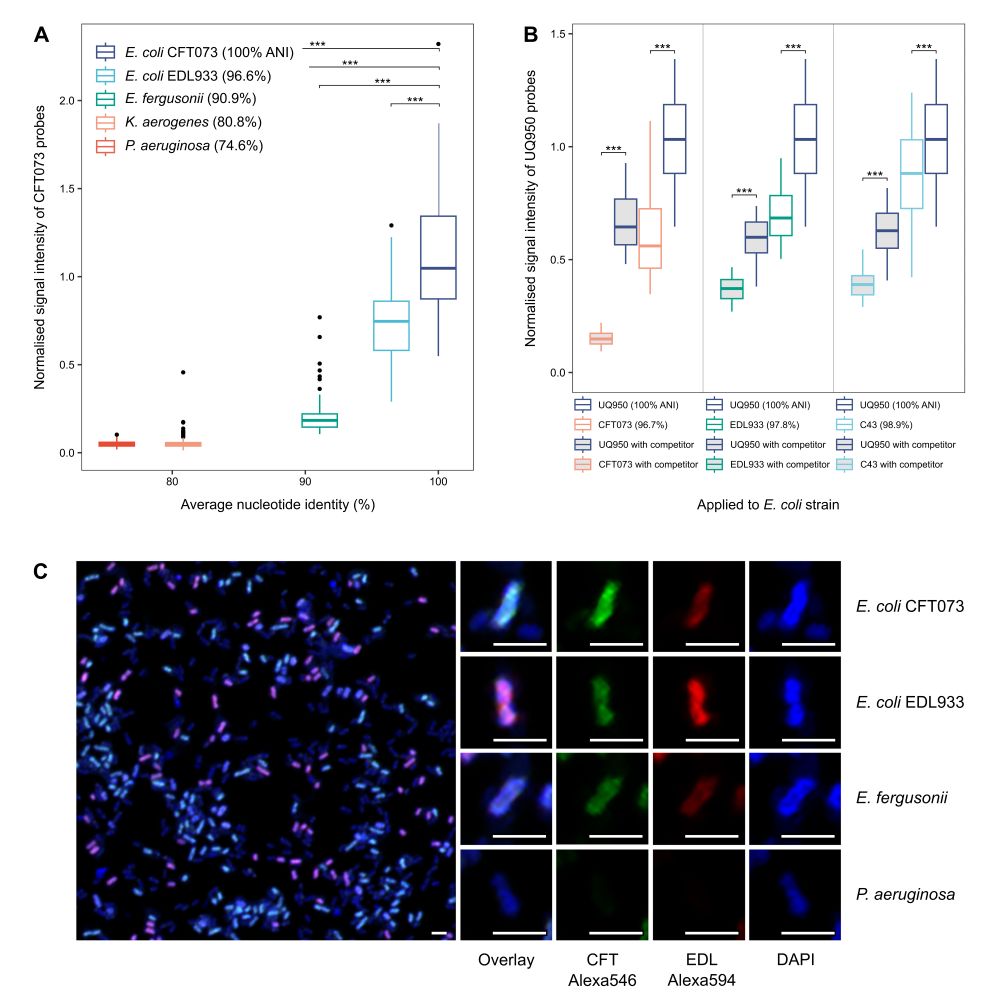
As a result, GenomeFISH can distinguish between strains with up to 99% ANI in mock communities and environmental samples.
08.07.2025 02:32 — 👍 2 🔁 1 💬 1 📌 0
By targeting the entire genome, GenomeFISH overcomes the inherent limitations of traditional FISH, circumventing the need for probe design and optimisation, while increasing sensitivity and specificity.
08.07.2025 02:32 — 👍 1 🔁 1 💬 2 📌 0
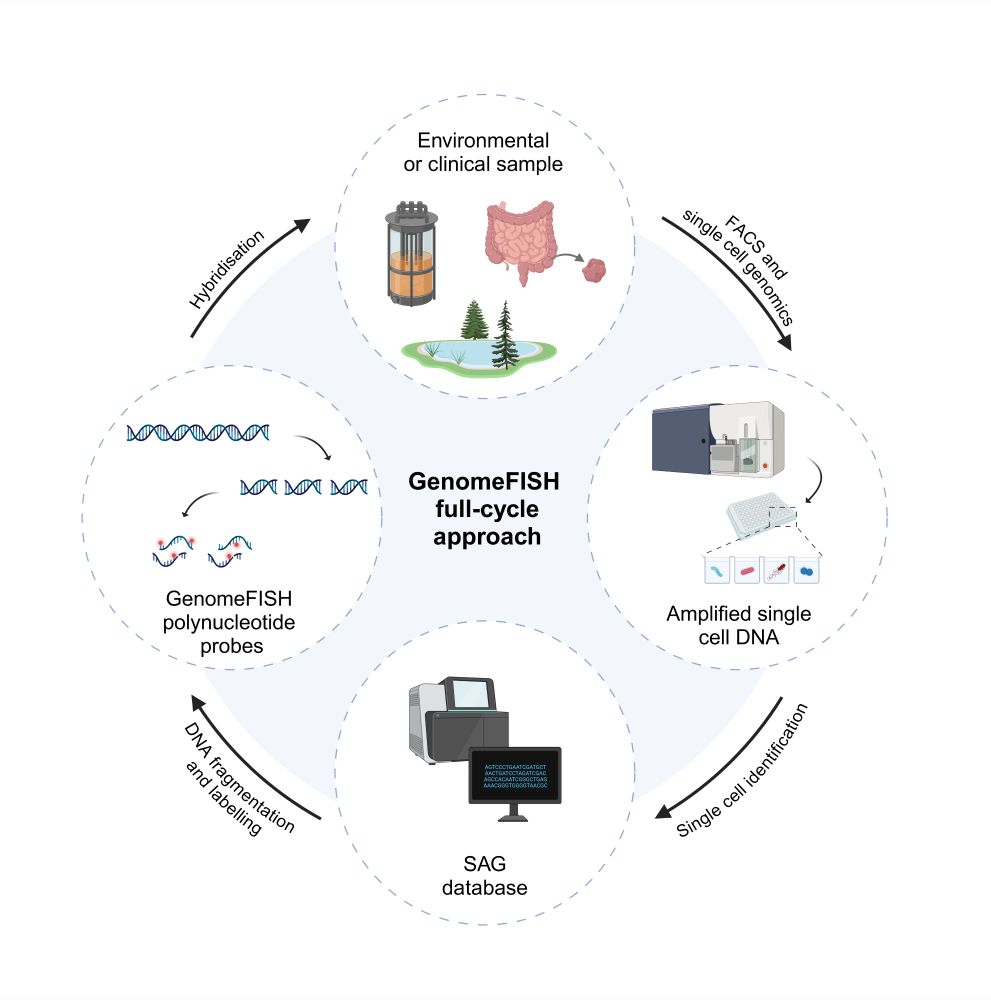
GenomeFISH probes can be rapidly generated from single cell genomes obtained from environmental and clinical samples, which are amplified, fragmented, fluorescently labelled, and hybridised to visualise the target microorganism in situ.
08.07.2025 02:31 — 👍 1 🔁 1 💬 1 📌 0
GenomeFISH is an innovative, genome-based FISH approach that couples high-throughput single-cell genomics with whole-genome hybridisation to visualise microbial communities.
08.07.2025 02:31 — 👍 2 🔁 1 💬 1 📌 0

GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities
Abstract. Fluorescence in situ hybridisation (FISH) is a powerful tool for visualising the spatial organisation of microbial communities. However, traditio
Very excited to share the first paper out of my Postdoc @CMR:
GenomeFISH: genome-based fluorescence in situ hybridisation for strain-level visualisation of microbial communities.
@sjmcilroy.bsky.social @jamesvolmer.bsky.social @benjwoodcroft.bsky.social
doi.org/10.1093/isme...
🧵1/7
08.07.2025 02:30 — 👍 63 🔁 27 💬 4 📌 2

Hello BrisJAMS Community :)
BrisJAMS returns this July with an Antimicrobial Resistance Special featuring three fantastic speakers.
📅 Wednesday, 23 July 2025
🕕 6:00–7:30 PM
📍 The Burrow, West End
🍽 Free food provided!
This free event is open to all in the microbiology community.
07.07.2025 06:39 — 👍 2 🔁 2 💬 1 📌 1
Lyrebird (phage profiling)
Documentation for SingleM
First code release of "SingleM for dsDNA phage"! Lyrebird scans metagenomic reads for marker genes to give a “phage community profile”. It detects many novel phages, many more than standard contig-centric methods. @benjwoodcroft.bsky.social @emerge-bii.bsky.social wwood.github.io/singlem/Lyrebird
16.05.2025 10:55 — 👍 12 🔁 8 💬 0 📌 1
Short-read metagenomic sequencing cannot recover genomes from many abundant marine prokaryotes due to high strain heterogeneity and platform-inherent GC bias (likely viruses, too), but Nanopore long reads can address this. A results thread on our recent preprint 🧵.
25.05.2025 10:27 — 👍 44 🔁 23 💬 1 📌 4
Please check out our recent preprint from @ace-uq.bsky.social researcher’s @stevenjrobbins.bsky.social, @xrefugee13.bsky.social, and former ACEr @pam-engelberts.bsky.social on their work generating the Great Barrier Reef Microbial Genomes Database (GBR-MGD).
21.05.2025 10:36 — 👍 6 🔁 2 💬 1 📌 0

The planktonic microbiome of the Great Barrier Reef
Large genome databases have markedly improved our understanding of marine microorganisms. Although these resources have focused on prokaryotes, genomes from many dominant marine lineages, such as Pela...
Very excited to present the Great Barrier Reef Microbial Genomes Database (GBR-MGD), a comprehensive DB of 1000s of high-quality prokaryote, virus, plasmid, and chromosome-level eukaryote MAGs using Nanopore long reads. Subthreads incoming. Please share widely. 🙂
www.biorxiv.org/content/10.1...
21.05.2025 07:34 — 👍 113 🔁 67 💬 2 📌 11
Postdoctoral researcher | Biotechnologist | Anaerobic microbiology | Methanotrophs | aSRB | LAB | Microbial Physiology group @cuwelte.bsky.social
CryoEM, Structural Cell Biology. Group Leader@University of Melbourne. #NHMRC Emerging Leadership Fellow. Formerly at @Caltech, @MRC_LMB.
Molecular microbial ecology & beneficial microbiomes & plant-microbe interactions. PhD & current postdoc at University of Copenhagen. Swim bike run
Publishing the best of biotech science and business. Find us on Twitter, Facebook & Instagram. Part of @natureportfolio.nature.com.
Official Bluesky account of the AHMB.
Expanding our access to the human microbiome
http://ahmb.com.au
Associate prof at Cal Poly Humboldt. Ecologist of microbes, tasted metagenomics once and now I want more. Spawned in Colombia, cumbia follows me everywhere I go.
09-13th September 2025, Vila do Conde, Portugal
1 phylum - 5 days - a global community
Website: www.worldspongecommunity.com
Donate: https://events.ciimar.up.pt/index.php/event/12th-world-sponge-conference-donations/
Located at the University of California-Berkeley at the Innovative Genomics Institute.
Purveyors of Microbial Ecology, Bioinformatics, & Nanogeoscience.
Reposts or likes≠endorsements.
https://www.banfieldlab.com/
Centre for Microbiology and Environmental Systems Science at @univie.ac.at, pioneering microbiome and environmental research for the benefit of the planet.
environmental microbiologist fascinated by the unseen majority | scientist at division of microbial ecology, university of vienna
Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) is an initiative to establish a standardised microbial taxonomy based on genome phylogeny.
Microbiologist :)
Studying bacterial pathogenesis, AMR & antimicrobial development.
Research fellow @ Institute for Molecular Bioscience, UQ
Previously Postdoc @Dunn School of Pathology, University of Oxford.
PhD @The Australian National University
Senior bioinformatician at Australian Centre for Ecogenomics (ACE).
Nature Microbiology publishes the latest research and commentary in all areas of microbiology.
https://www.nature.com/nmicrobiol/
Exploring the wonders of microbiology with reviews and commentaries from across the field. Brought to you by your curious editors! 🔬✨
https://nature.com/nrmicro/
Infectious Disease Epidemiologist - Nontuberculous mycobacteria: Research Fellow at the University of Queensland | Microbial Genomics | Also interested in evolution, pathogenesis and #AMR of clinically relevant bacterial pathogens | She/her | Cat lover
Molecular Ecologist | AIMS@JCU Alumni
Research Fellow at IMB-UQ, Bacteriologist. Bacterial cell envelope enthusiast. Astrophotography lover. Dad of two. Personal Acc, Apoliticism.













