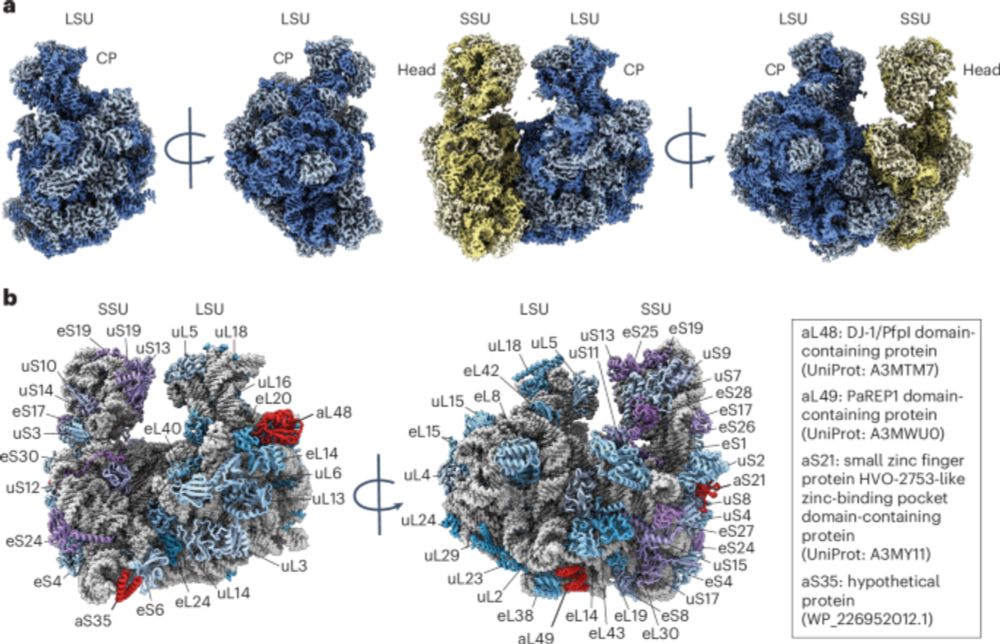
🌀 We solved the structure for T. denticola periplasmic flagella!
I'm very happy to finally showcase this work. A huge collaborative effort between myself, @bindusmitapaul.bsky.social, @debnathghosal.bsky.social, Jack Kim, @banfieldlab.bsky.social, Eric Reynolds and the Chris Fenno lab.
🦠 Enjoy!
06.02.2026 04:23 — 👍 5 🔁 3 💬 0 📌 0

Today in Nature Communications, a team of IGI researchers from The Banfield Lab and Pam Ronald's labs uncover a new way to reduce methane emissions from rice by influencing the activity of rice paddy soil #microbes. Read more: https://ow.ly/45j150Y3WsI
26.01.2026 21:05 — 👍 15 🔁 7 💬 1 📌 0
🚨 New pre-print alert!
Jumbo circular extrachromosomal elements of methane-oxidizing archaea with variably extensive metabolic and defense gene repertoires
www.biorxiv.org/content/10.6...
23.01.2026 01:21 — 👍 13 🔁 7 💬 0 📌 0
Excited to share our discovery of archaeal circular, jumbo extrachromosomal elements (up to ~535 kb genomes). One related, 409-kb genome is integrated in CH4-eating Methanoperedens, representing the largest integrative element in Archaea so far! Curious about what they are doing? See the paper
23.01.2026 00:58 — 👍 18 🔁 12 💬 1 📌 1

A new preprint from The Banfield Lab looks at mobile genetic elements with single-cell precision. Read here: https://ow.ly/hFBX50Y0nQA
#microbiology
21.01.2026 16:05 — 👍 10 🔁 3 💬 1 📌 0

A new preprint on expanding the genetic code, from IGI's Jill Banfield, Peter Penev, and collaborators across the country: https://ow.ly/HiYO50XVNXR
15.01.2026 17:05 — 👍 14 🔁 4 💬 0 📌 0

Postdoc - Microbiome - Innovative Genomics Institute
University of California, Berkeley is hiring. Apply now!
🚨 Our Lab is HIRING! If you are interested in characterizing model microbiomes using genome-resolved methods, statistical/metabolic modeling, and/or machine learning please submit an application here: aprecruit.berkeley.edu/JPF05234
#postdoc #sciencejobs #biology #microbiome
05.01.2026 18:19 — 👍 12 🔁 9 💬 1 📌 0
✨New preprint!
🧵1/4 Excited to share our work on AI-guided design of minimal RNA-guided nucleases. Amazing work by @petrskopintsev.bsky.social @isabelesain.bsky.social @evandeturk.bsky.social et al!
Multi-lab collaboration @banfieldlab.bsky.social @jhdcate.bsky.social @jacobsenucla.bsky.social🧬
🔗👇
09.12.2025 07:52 — 👍 98 🔁 47 💬 1 📌 8

illustration of petri dishes growing microbes
Out today in Science Magazine — First author Veronika Kivenson and PIs Jill Banfield (The Banfield Lab) and Alanna Schepartz team up to reveal a new genetic code in #archaea, with implications for #methane and #climate, and #bioengineering! Learn more: https://ow.ly/Kuem50Xurh0
08.12.2025 15:50 — 👍 15 🔁 6 💬 0 📌 0
Incredible science coming out from our lab. Congrats @kivenson.bsky.social 🎉
20.11.2025 19:53 — 👍 13 🔁 5 💬 0 📌 0
Fresh from the press! Congrats @diamondlab.bsky.social
19.11.2025 23:03 — 👍 4 🔁 0 💬 0 📌 0
We tried to contact researchers at Sichuan Agricultural University who may have deposited this data (PRJNA601687) with no reply. Please let us know if you have any information.
06.10.2025 23:42 — 👍 1 🔁 0 💬 0 📌 0
Newest preprint from the group on megaplasmids in E. coli led by @assemblerag.bsky.social. In addition to our findings in metagenomic datasets, we identified an RNA-seq project from a giant panda gut sample with evidence of megaplasmids. We would like to include the data creators in our study.
06.10.2025 23:42 — 👍 9 🔁 3 💬 1 📌 0

The 2025 Banfield Retreat was fantastic 🦠 so many talented scientists in one place is a sure way to come up with some great ideas!
@banfieldlab.bsky.social
01.05.2025 18:06 — 👍 4 🔁 2 💬 0 📌 0
🚨 Fresh from the press! We created and analyzed over 100 in vitro cyanobacterial consortia using well-characterized model cyanobacterial hosts to better understand how cyanobacteria recruit and interact with their microbiomes.
Check it out: doi.org/10.1093/isme...
11.07.2025 13:07 — 👍 45 🔁 24 💬 3 📌 4
Many of you gave me so much encouragement when I posted here earlier about my 10 year long passion project to make a microbial map of the ocean. Pinch me because I can't believe I get to update you that today that research was published in the journal SCIENCE! @science.org
10.07.2025 19:29 — 👍 18 🔁 10 💬 4 📌 1
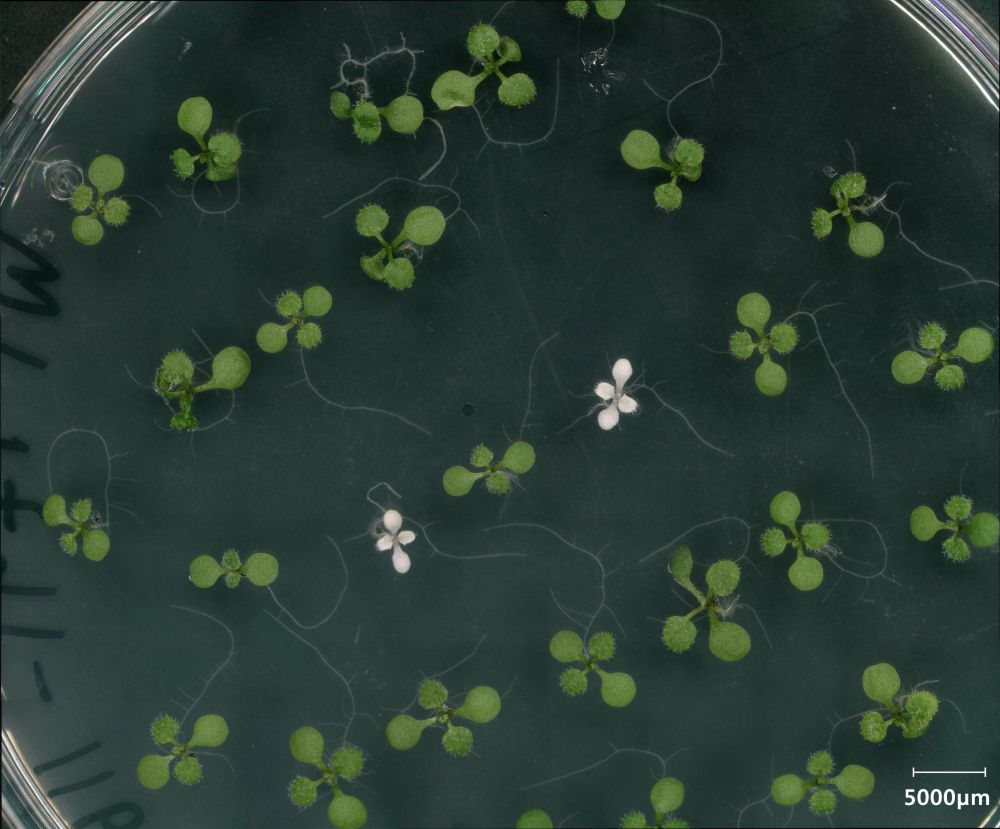
Tiny CRISPR Tool Opens the Door to Faster, Simpler Plant Genome Editing
Tiny CRISPR Tool Opens the Door to Faster, Simpler Plant Genome Editing - Innovative Genomics Institute (IGI)
In a recent paper, IGI's @jenniferdoudna.bsky.social @doudna-lab.bsky.social and Jill Banfield @banfieldlab.bsky.social collaborate with UCLA's @jacobsenucla.bsky.social to create a new, tiny #CRISPR tool for quicker, easier plant genome editing! Read more in this Q&A with Steve: ow.ly/6Szs50VGElf
31.07.2025 23:17 — 👍 9 🔁 4 💬 0 📌 0
Most plasmids described in E. coli are small compared to the megaplasmids we identified here! Check out the preprint if you want to learn about these mysterious large elements and their potential functions 🧬. I’m very grateful to have had the opportunity to work on this in @banfieldlab.bsky.social
01.10.2025 05:10 — 👍 34 🔁 19 💬 2 📌 1
Excited to share new research led by our very own @lingdong-shi.bsky.social showing that many archaeal 23S rRNAs are circularized in active ribosomes. Check out the preprint below.
29.04.2025 21:34 — 👍 31 🔁 14 💬 0 📌 0
Genomes from long-read metagenomic assemblies contain rampant errors, highlighting the pressing need for stricter evaluation methods in long-read assembly algorithms. Read more in our paper with the Eren group. @floriantrigodet.bsky.social @merenbey.bsky.social
28.04.2025 21:29 — 👍 42 🔁 16 💬 0 📌 2
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
Beyond Known Biology
basecamp-research.com
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
Rohan Sachdeva is a computational microbiologist who uses (meta)genomics and an (un)healthy dose of caffeine at the Innovative Genomics Institute.
A graduate student-run publication highlighting scientific advancements in climate, technology, biology, physics, chemistry and engineering.
Read Online -> https://berkeleysciencereview.com/
Join Our Team -> https://linktr.ee/berkeleyscirev?fbclid=PAZ
Publishing the best of biotech science and business. Find us on Twitter, Facebook & Instagram. Part of @natureportfolio.nature.com.
News from the Rubin Lab 🧬🧫 We conduct DNA-level editing of microbial communities for understanding and control. Managed by lab members.
@innovativegenomics.bsky.social @ucberkeleyofficial.bsky.social 🔬🌁
🔗 therubinlab.org
Yet another microbial bioinformatician, group leader, dad
github.com/wwood https://research.qut.edu.au/cmr/team/ben-woodcroft/
UC Davis rice geneticist & co-author of Tomorrow's Table: Organic Farming Genetics & the Future of Food. TED talk. My lab studies genetics for crop resilience & #plant-microbe interactions. Volunteer Ranger USFS. Nature photography
Lifelong learner exploring microbial ecology, microbiomes, microbe–microbe and host–microbe interactions. @ GEOMAR Helmholtz Centre for Ocean Research | @ Kiel University
molecular and cellular bio PhD candidate in the Cress and Doudna labs @UC Berkeley
Assistant Professor of Environmental Microbiology at ETH Zurich | https://envmicrobiol.ethz.ch/
The International Society for Microbial Ecology is a non-profit association and owner of the ISME Journal and ISME Communications.
Next symposium #ISME20 in Auckland, New Zealand in August 2026.
The Diamond Lab | Innovative Genomics Institute @ UC Berkeley | Metagenomics | Microbiome Systems Ecology and Modeling | AI for Microbes | Environmental MGEs | Health and Climate Change
https://www.diamondlab.bio/
@innovativegenomics.bsky.social
biophysics phd student in the banfield lab @ uc berkeley | protein structures, evolution, mobile genetic elements, bacterial immune systems
🕸️ kinseylong.github.io
Postdoc at IGI @ UC Berkeley |Metagenomics | Bioinformatics | Algorithm | Human genome
chem bio PhD student in the Doudna Lab @Berkeley
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing