Remember the slogan projects used to have: "Made with ❤️ by XYZ"?
Soon we’ll start seeing: "Made by humans for humans"
27.01.2026 08:40 — 👍 0 🔁 0 💬 0 📌 0

course schedule as a table. Available at the link in the post.
I'm teaching Statistical Rethinking again starting Jan 2026. This time with live lectures, divided into Beginner and Experienced sections. Will be a lot more work for me, but I hope much better for students.
I will record lectures & all will be found at this link: github.com/rmcelreath/s...
09.12.2025 13:58 — 👍 659 🔁 235 💬 12 📌 20

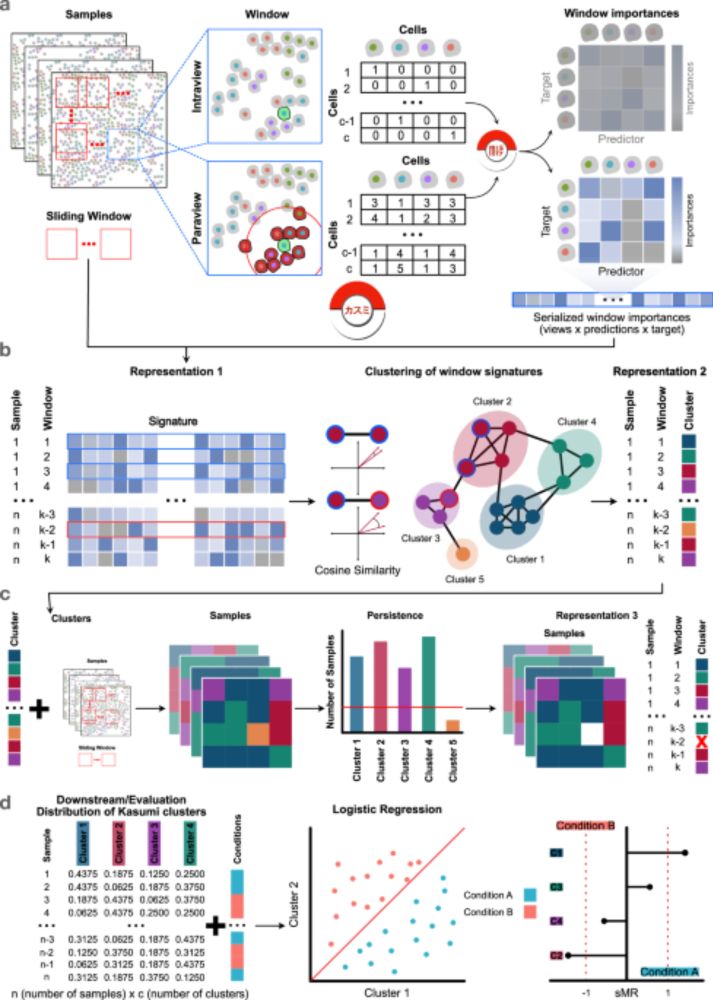
Unifying multi-sample network inference from prior knowledge and omics data with CORNETO
Mixed-integer optimization Mixed-integer programming is a type of constrained optimization problem that involves decision variables that can be both integer and continuous. A mixed-integer programming problem can be defined as follows: $$\begin{array}{ll}\mathop{\min }\limits_{{\bf{x}}\in {{\mathbb{R}}}^{n},\,{\bf{y}}\in {{\mathbb{Z}}}^{m}}&f({\bf{x}},{\bf{y}})\\ \,\text{subject to}\,&{g}_{i}({\bf{x}},{\bf{y}})\le 0,\quad i=1,2,\ldots ,k,\\...
Unifying multi-sample network inference from prior knowledge and omics data with CORNETO
->Nature | #Data | More info from EcoSearch
23.07.2025 05:23 — 👍 1 🔁 1 💬 0 📌 0

🚨 New preprint
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
19.09.2025 07:45 — 👍 30 🔁 7 💬 1 📌 1
ScAPE: A lightweight multitask learning baseline method to predict transcriptomic responses to perturbations https://www.biorxiv.org/content/10.1101/2025.09.08.674873v1
09.09.2025 19:47 — 👍 2 🔁 2 💬 0 📌 0

We present our MetaProViz #Rpackage for #metabolomics analysis & prior knowledge integration to generate mechanistic hypotheses on how metabolic changes affect metabolite classes, pathways & environment interaction
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
25.08.2025 07:07 — 👍 25 🔁 13 💬 2 📌 0

EMBO Practical Course. Causality in biomedicine: going beyond associations. 4 - 9 October 2026. Hinxton, UK. EMBO Practical Course and EMBL-EBI logos attached.
New course announced!
We're thrilled to be hosting the @embo.org Practical Course 'Causality in biomedicine: going beyond associations' from 4 – 9 October 2026.
Register your interest and be the first to hear when the course opens for applications: www.ebi.ac.uk/training/eve...
12.08.2025 11:50 — 👍 9 🔁 5 💬 1 📌 1
New course “EMBO Causality in Biomedicine”: We have organised the first EMBO course in *causal* stats/ML methods for quantitative biomedicine. @sjoerdvbeentjes.bsky.social @nimahejazi.org @pablormier.bsky.social @DariaSokolova @CarolineUhler
Very much looking forward to teaching and discussing!
12.08.2025 14:03 — 👍 10 🔁 7 💬 0 📌 0
This project has been in the making for quite some time. CORNETO not only integrates key concepts and methodologies in biological network inference, but also introduces a novel framework for multi-condition analysis. Congrats to the team, and especially to @pablormier.bsky.social for leading this.
24.07.2025 07:51 — 👍 7 🔁 3 💬 0 📌 0
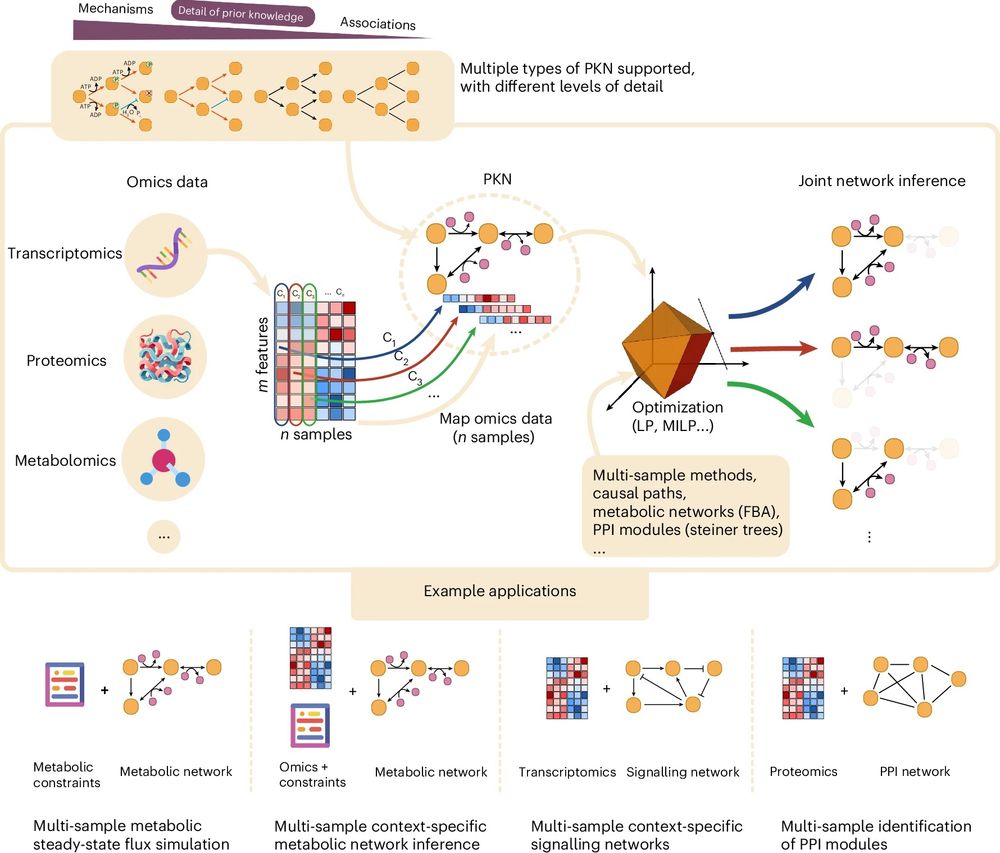
🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is published in peer reviewed form
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
22.07.2025 15:23 — 👍 61 🔁 29 💬 1 📌 1
CORNETO: machine learning to decode complex omics data
New tool combines biological knowledge with machine learning to help researchers extract meaningful insights from complex omics data.
How can we find out what’s really going on inside cells when we’re generating so much complex data?
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
22.07.2025 09:20 — 👍 38 🔁 10 💬 1 📌 0
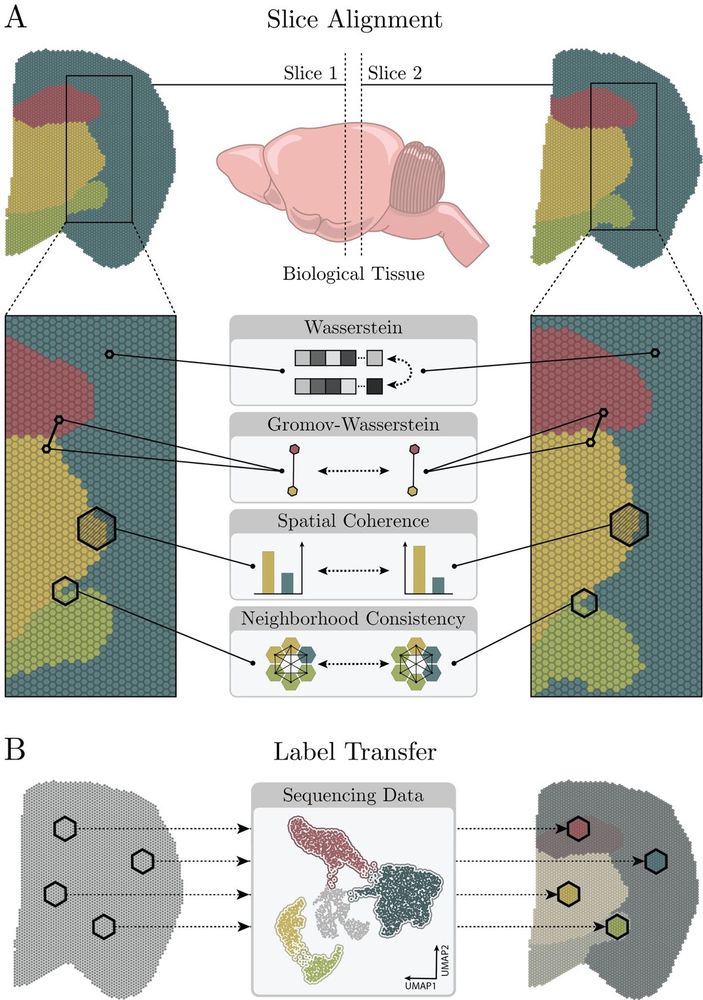
🚨 New preprint: Topography Aware Optimal Transport for Alignment of Spatial Omics Data
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
22.04.2025 09:37 — 👍 31 🔁 17 💬 1 📌 1

Pretraining virtual cells is useless
the way we do it now
Turns out the way we usually pre-train foundational cell models adds very little information to the system - definitely not enough to make drug effect predictions work. Not what I've expected.
#virtualcells #foundationalmodels #compbio
blog.turbine.ai/p/pretrainin...
16.04.2025 17:03 — 👍 2 🔁 2 💬 0 📌 0
📄 Update on our preprint about Gene Regulatory Net (GRN) benchmarking 📄
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇
14.03.2025 10:34 — 👍 50 🔁 18 💬 2 📌 0
No need to feel bad! Also, congratulations on your work, and best wishes to Sakana!
17.03.2025 11:42 — 👍 1 🔁 0 💬 0 📌 0
Is someone feeling jealous? What's the beef here? 🥩
16.03.2025 22:58 — 👍 0 🔁 0 💬 0 📌 0


Haha, Peyman Milanfar blocked me within milliseconds after I liked a reply of somebody else defending @hardmaru.bsky.social and Sakana AI against his attacks.
Fastest block ever!
16.03.2025 22:55 — 👍 1 🔁 0 💬 2 📌 0
YouTube video by Stanford Online
Stanford EE364A Convex Optimization I Stephen Boyd I 2023 I Lecture 1
Stephen Boyd's reaction when a student says they're using Genetic Algorithms for optimization is priceless 😂
youtu.be/kV1ru-Inzl4?...
16.03.2025 17:15 — 👍 2 🔁 1 💬 0 📌 0
In many countries we're seeing voters that have never known anything other than stability and a certain level of competence voting for anti- system candidates because they have convinced themselves of two things: things right now are awful; change will only be for the better.
03.02.2025 08:27 — 👍 242 🔁 61 💬 1 📌 4
This is also why vaccine-denial is so prevalent. People don’t have a memory of what mass death from smallpox looked like, or post-poli disability.
24.02.2025 10:50 — 👍 68 🔁 7 💬 4 📌 0
No AGI until LLMs can reliably produce useful LaTeX. Haven’t seen one that truly delivers.
We need a LLM LaTeX benchmark!
20.02.2025 18:34 — 👍 2 🔁 0 💬 0 📌 0

"LLaDA: Large Language Diffusion Models" Nie et al.
Just read this fascinating paper.
Scaled up Masked Diffusion Language Models to 8B params, and show that it can match #LLMs (including Llama 3) while solving some key limitations!
Let's dive in... 🧵
(1/8)
#genai
18.02.2025 15:05 — 👍 1 🔁 1 💬 1 📌 0
Overview - Health - ELSA Benchmarks Platform
We are delighted to announce The Health Privacy Challenge, an interactive opportunity for advancement at the intersection of computational biology and privacy research, brought to you as a part of CAMDA Conference at #ISMB/ECCB2025 🫐🍅
Register to participate: benchmarks.elsa-ai.eu?ch=4
15.01.2025 09:27 — 👍 9 🔁 8 💬 1 📌 1
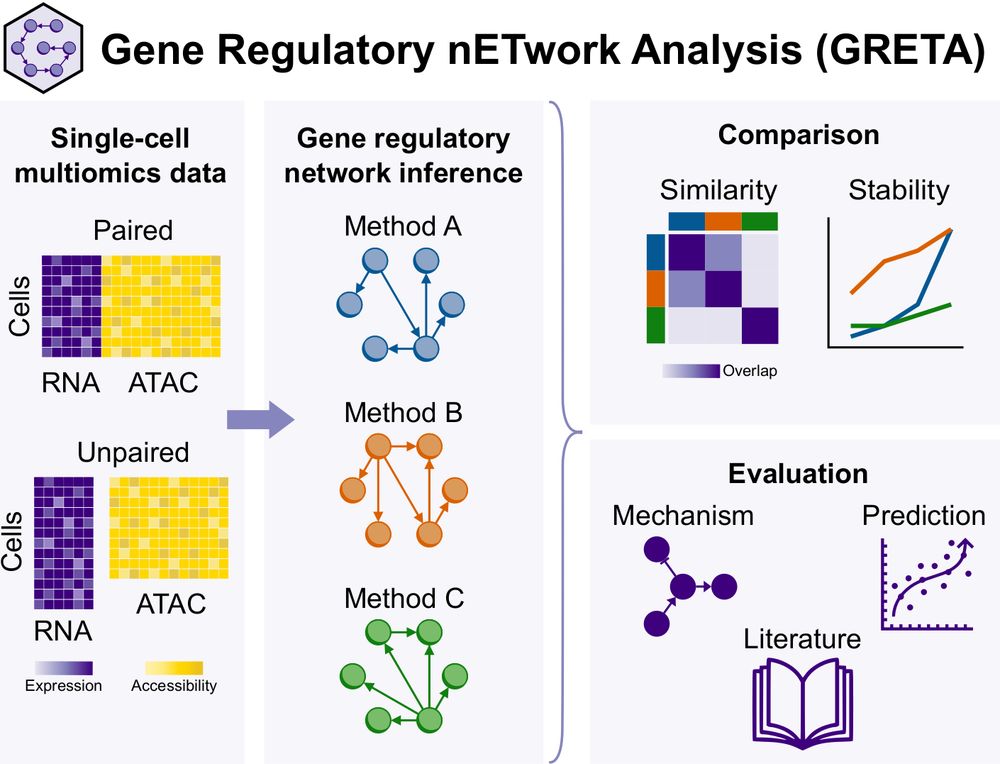
GRETA graphical abstract
We present Gene Regulatory nETwork Analsyis (GRETA), a framework to infer, compare and evaluate gene regulatory networks #GRNs. With it, we have benchmarked multimodal and unimodal GRN inference methods. Check the results here 👇
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
23.12.2024 08:43 — 👍 128 🔁 46 💬 1 📌 5
The final chapter of my PhD thesis is now out! 🎉 We compared the latest gene regulatory network (#GRN) inference methods for #single-cell multimodal datasets and evaluated their performance across various tasks. Hard to believe this journey started in March 2021 and has finally reached this point 😅🥳
23.12.2024 08:49 — 👍 31 🔁 8 💬 0 📌 1
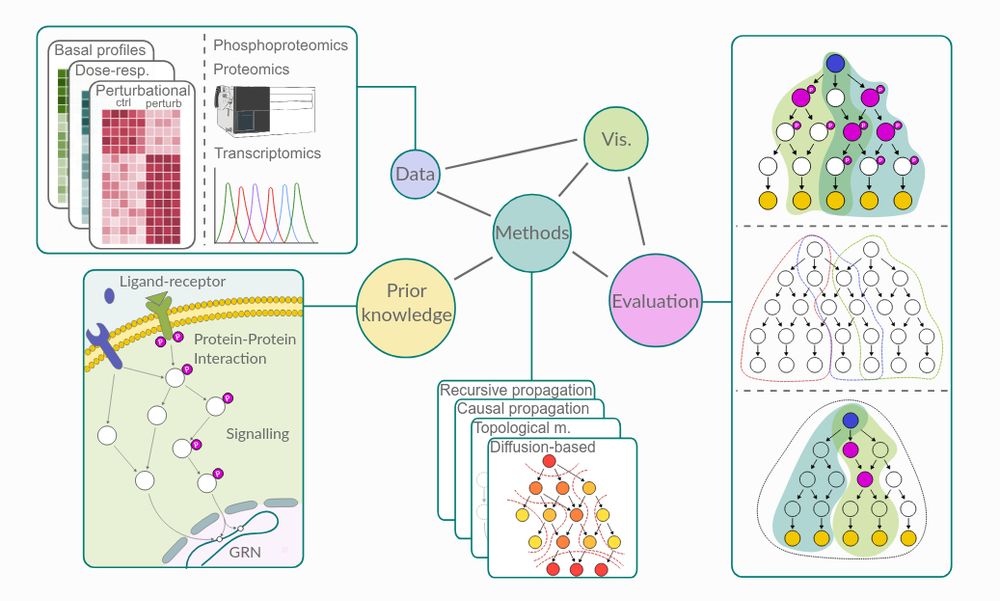
We present NetworkCommons, a unified platform 🪐 for network biology, providing access to omics data, knowledge, and contextualization methods, all with a consistent API 👇🧵
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
26.11.2024 09:44 — 👍 96 🔁 32 💬 2 📌 4

How cute is this little guy? 😍
22.11.2024 21:11 — 👍 3 🔁 0 💬 0 📌 0
Computational biologist using omics data (imaging, gene expression) to investigate the impacts of environmental contaminants on biological systems
Group Leader @EMBL-EBI (www.ewaldlab.org)
Learning to optimize | graph neural networks | AI 4 science
PhD student at Uppsala University, incoming research intern at Microsoft
PhD student at Fraticelli lab @irbbarcelona.org
I try to convert coffee into code and ideas for uncovering biological mysteries.
Some interests: single-cell, lineage tracing, computational biology, cellular variability, premalignancy, resistance...
Econ professor at Michigan ● Senior fellow, Brookings ● Intro econ textbook author ● Think Like An Economist podcast ● An economist willing to admit that the glass really is half full ● Find me: https://linktr.ee/justinwolfers
Full Professor of Computational Statistics at TU Dortmund University
Scientist | Statistician | Bayesian | Author of brms | Member of the Stan and BayesFlow development teams
Website: https://paulbuerkner.com
Opinions are my own
Academy Professor in computational Bayesian modeling at Aalto University, Finland. Bayesian Data Analysis 3rd ed, Regression and Other Stories, and Active Statistics co-author. #mcmc_stan and #arviz developer.
Web page https://users.aalto.fi/~ave/
Zealous modeler. Annoying statistician. Reluctant geometer. Support my writing at http://patreon.com/betanalpha. He/him.
human being | assoc prof in #ML #AI #Edinburgh | PI of #APRIL | #reliable #probabilistic #models #tractable #generative #neuro #symbolic | heretical empiricist | he/him
👉 https://april-tools.github.io
Decision-making under uncertainty, machine learning theory, artificial intelligence · anti-ideological · Assistant Research Professor, Cornell
https://avt.im/ · https://scholar.google.com/citations?user=EGKYdiwAAAAJ&sortby=pubdate
AI, sociotechnical systems, social purpose. Research director at Google DeepMind. Cofounder and Chair at Deep Learning Indaba. FAccT2025 co-program chair. shakirm.com
Assistant Professor of Machine Learning
Generative AI, Uncertainty Quantification, AI4Science
Amsterdam Machine Learning Lab, University of Amsterdam
https://naesseth.github.io
Theory & practice of probabilistic programming. Current: MIT Probabilistic Computing Project; Fall '25: Incoming Asst. Prof. at Yale CS
Associate Professor (UHD) at the University of Amsterdam. Probabilistic methods, deep learning, and their applications in science in engineering.
#AI4Science #CompNeuro #NeuroAI #SBI
www.mackelab.org @mackelab.bsky.social
· Prof Uni Tuebingen ML4Science BCCN tue.ai
· Adjunct MPI IS · Fellow ellis.eu
· currently hiring postdocs and PhD students
· sometimes goes for a run
ELLIS PhD student at the University of Edinburgh
https://lenazellinger.github.io/
Assistant Professor, University of Toronto.
Junior Research Fellow, Trinity College, Cambridge.
AI Fellow, Georgetown University.
Probabilistic Machine Learning, AI Safety & AI Governance.
Prev: Oxford, Yale, UC Berkeley, NYU.
https://timrudner.com
Machine Learning Researcher
https://alexalemi.com
https://blog.alexalemi.com
Assoc. Prof. of Machine & Human Intelligence | Univ. Helsinki & Finnish Centre for AI (FCAI) | Bayesian ML & probabilistic modeling | https://lacerbi.github.io/