
Want to learn about the theoretical aspects of proteins/peptides analysis by mass spectrometry?
Then join the #EMBLProteomics course where you will get hands-on experience in sample preparation and perform tandem mass tag labelling.
Apply by 3 March:https://s.embl.org/sdp26-01-bl 🔬💻
03.12.2025 09:32 — 👍 4 🔁 3 💬 0 📌 0
Finally out! 🤩 Check out our HT- PELSA for high throughput screening!
05.11.2025 16:28 — 👍 16 🔁 6 💬 1 📌 1
Happy to see our HT-PELSA paper now published in @natsmb.nature.com 🎊 Big thanks for the constructive review process! 📖Read the manuscript here (www.nature.com/articles/s41...) & check the thread for additional information ⬇️
05.11.2025 10:15 — 👍 36 🔁 14 💬 0 📌 1
CORNETO: machine learning to decode complex omics data
New tool combines biological knowledge with machine learning to help researchers extract meaningful insights from complex omics data.
How can we find out what’s really going on inside cells when we’re generating so much complex data?
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
22.07.2025 09:20 — 👍 38 🔁 10 💬 1 📌 0
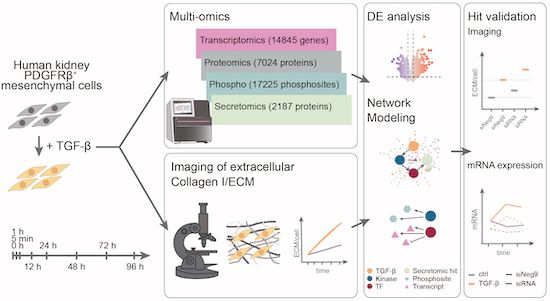
integrated temporally resolved transcriptomic, proteomic, and secretomic profiling of human kidney mesenchymal cells from @savitski-lab.bsky.social @saezlab.bsky.social @miraburtscher.bsky.social identify dynamic changes upon TGF-beta stimulation ➡️ www.embopress.org/doi/full/10....
03.07.2025 12:30 — 👍 4 🔁 3 💬 0 📌 0
Big congrats to everyone involved - it was a blast! 🎊 If you're interested in #multiomics integration or #kidneyfibrosis, check it out! ⬇️
04.07.2025 07:18 — 👍 4 🔁 1 💬 0 📌 0
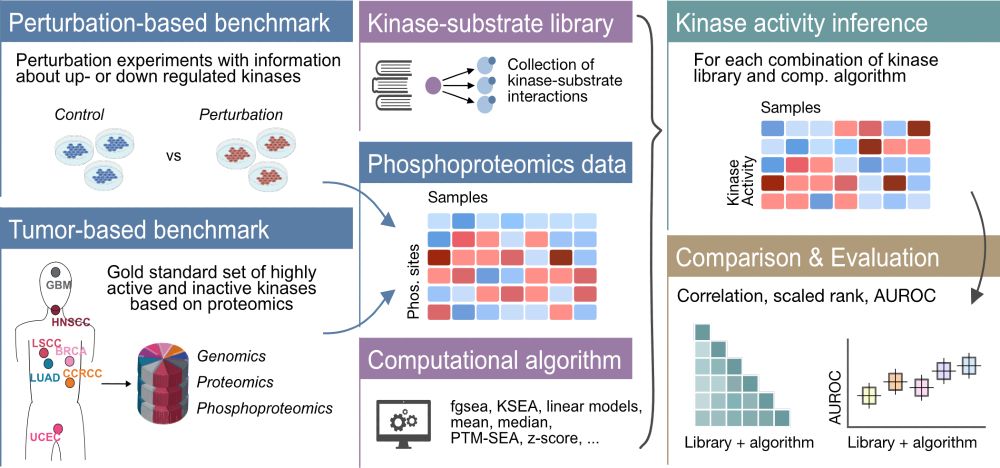
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab @bcmhouston.bsky.social, is now out @natcomms.nature.com 🔬 tinyurl.com/4twuc6z4
23.05.2025 07:37 — 👍 47 🔁 20 💬 1 📌 2
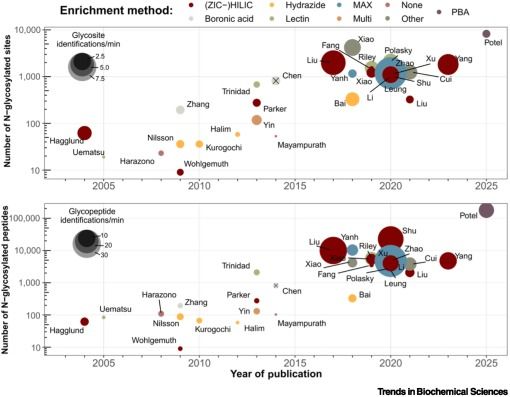
Online now - the Spotlight "Accelerating the stride toward functional #glycoproteomics" from @timveth.bsky.social and @nmriley.bsky.social.
#Glycoproteins #Glycoforms #Glycoprofiling #DQGlyco #Glycotime
authors.elsevier.com/a/1l3k53S6Gf...
09.05.2025 13:41 — 👍 6 🔁 5 💬 1 📌 1
Excited to share our new paper! If you’re interested in glycoproteomics, be sure to check it out 👇
Huge kudos to all coauthors, especially the amazing glycoteam: Clement, @miraburtscher.bsky.social, Isabelle, and Misha (@savitski-lab.bsky.social)!
10.02.2025 12:45 — 👍 9 🔁 5 💬 0 📌 0
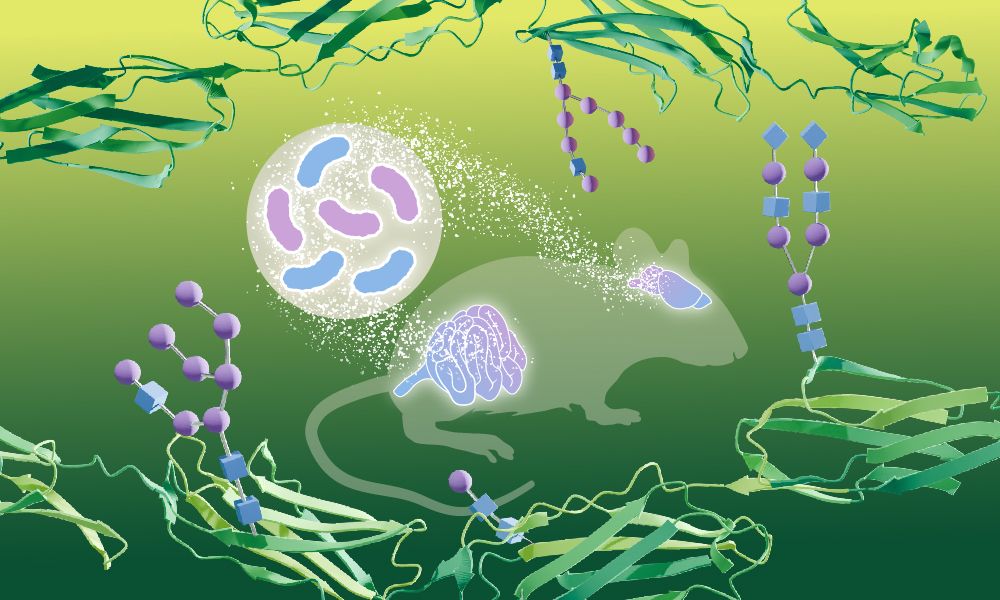
Bacteria can influence how sugars modify proteins in the brain – shown for the first time by EMBL researchers.
In their study, the scientists describe a new method to study glycosylation systematically & quantitatively, leading to new biological insights. 🧪🧠📈
www.embl.org/news/science...
10.02.2025 10:17 — 👍 46 🔁 13 💬 0 📌 0
Excited to see this huge piece of work out 🎉 Congrats to everyone involved! If you ever wanted to read a 8in1 paper grab a cup of tea and enjoy 📖
10.02.2025 10:32 — 👍 10 🔁 3 💬 0 📌 0
Science and fun from the lab of Tony Hyman at the MPI-CBG @mpicbg.bsky.social. Tweets by Hymanlab members.
https://hymanlab.org/
We love #ubiquitin. Lab researching ubiquitination in health and disease @WEHI-research. Posts are by Komander lab members.
Professor and Head of the Division of Molecular Therapeutics in MCB, UC Berkeley
Founder, Nurix and Lyterian Therapeutics
Excited about stress responses, ubiquitin, proteasome, targeted protein degradation, drug discovery
Molecular machines, Proteostasis, Ubiquitin Signaling, Cell Autonomous Defense, Protein Disorders and Team Nematode
@IMP Vienna
www.imp.ac.at/groups/tim-clausen/
👨🔬 Predoctoral Fellow at @EMBL.org in the @borklab.bsky.social.
💻🧬 Using data science to analyze big bio-data.
🇪🇺 Part of the @embltrec.bsky.social expedition and interested in 🦠 (metaG)
🇲🇽 Staff Scientist at EMBL-EBI (@ebi.embl.org and @saezlab.bsky.social) studying the molecular biology of multicellular disease processes, cellular cooperation and its parallels to ecology and social systems
Scientist at @saezlab.bsky.social, Heidelberg University. EMBL-EBI Visitor. Mathematical modeling / ML & AI for biological systems
https://pablormier.github.io/
Associate professor at ETH Zurich, studying the cellular consequences of genetic variation. Affiliated with the Swiss Institute of Bioinformatics and a part of the LOOP Zurich.
⚛️ Postdoc in Computational Biomedicine @saezlab.bsky.social @Heidelberg
🧪 Facinated by Cancer Metabolism, Gene Regulation & Multiomics
🎓 Previously @frezzalab.bsky.social @Cambridge_Uni and @Wuerzburg_Uni
🧫 Love to paint Scienceart
PhD student @EMBL in the Krebs Lab
Check out Microscopy Nodes for handling microscopy data in Blender! Now loading .tif and OME-Zarr with big volume support :)
she/her | IPA: ˈafkə ɡrɔs
PhD candidate at saezlab.bsky.social in computational biomedicine 🔬🧬
Die 1386 gegründete Ruperto Carola ist eine der forschungsstärksten in Europa.
#OpenAccess @embopress.org journal publishing pioneering research in systems & synthetic biology, systems medicine
Postdoc in Savitski lab, EMBL, Heidelberg. Interested in functional proteomics, microbiology, metabolism, and drug target discovery.
PhD student in the Savitski Team @EMBL Heidelberg. Alum Berezovski lab and JLHMS facility @uOttawa, @TUDarmstadt. Handball and Tennis and more coming
PhD at Schapiro lab in Heidelberg | Spatial biology | Bioinformatics | Tissue organization
PhD student in the Schulman Department at Max Planck Institute of Biochemistry 👩🏽🔬
BIF PhD Fellow 🧬
Interested in crosstalk between E3 Ligases and Metabolism






