The Weill Cancer Hub is fostering groundbreaking @ucsanfrancisco.bsky.social - @stanford-cancer.bsky.social collaborations to develop transformative cancer therapies. I’m grateful to co-lead one of the selected teams with @mfgrp.bsky.social to advance next-generation in vivo CAR T cell therapy!
25.07.2025 06:07 — 👍 7 🔁 0 💬 0 📌 0

Gift Launches $200M Initiative for the Weill Cancer Hub West
A $100 million matching grant from the Weill Family Foundation is bringing together two leading cancer centers to launch the Weill Cancer Hub West — an innovative collaboration among some of the natio...
Backed by a $100M gift from Joan and Sandy Weill, UCSF and @stanfordmedicine.bsky.social @stanford-cancer.bsky.social are launching Weill Cancer Hub West, a $200M initiative in team science to accelerate cancer research and improve care over the next decade.
tiny.ucsf.edu/rZIspe
23.07.2025 15:40 — 👍 6 🔁 3 💬 0 📌 2
Thank you for your amazing work at MSK, you inspired a generation whole of junior scientists and helped so many of us transitioning to our dream jobs!
01.03.2025 02:33 — 👍 1 🔁 0 💬 1 📌 0
10/ This was an awesome collab with the
@brshy.bsky.social Lab and led by Chris Chang. A huge shoutout to our amazing teams, collaborators and funders for making this possible!
@ucsfcancer.bsky.social @gladstoneinst.bsky.social @parkerici.bsky.social @pewhealth.bsky.social
05.02.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0
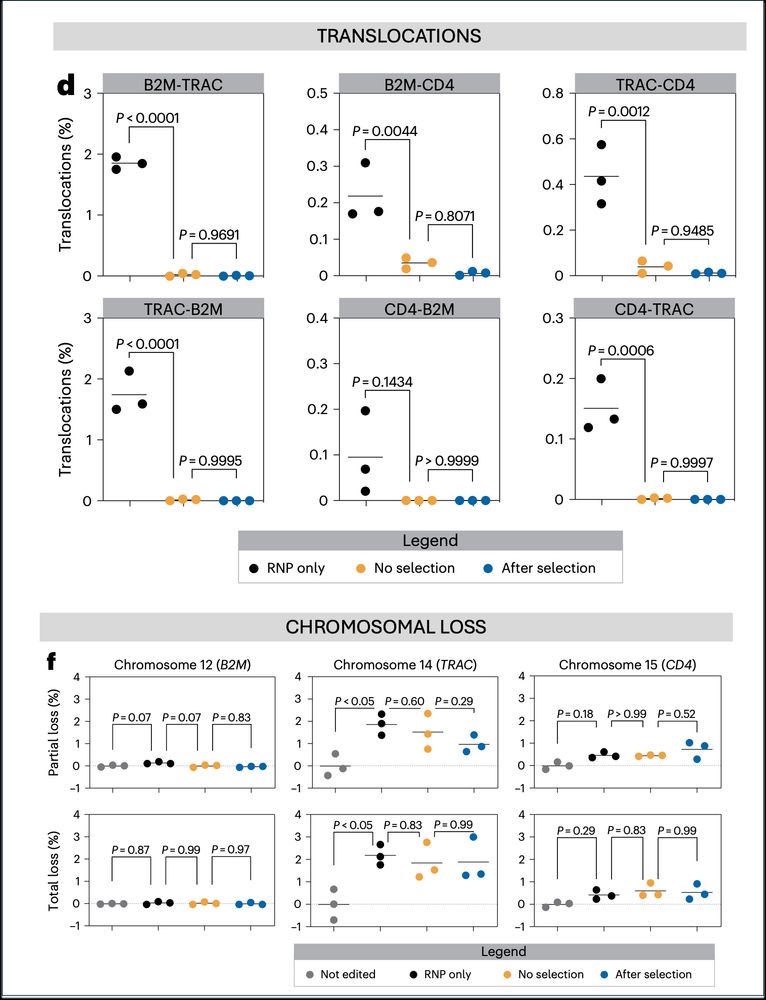
9/ Importantly, our optimized protocol did not lead to an increase of chromosomal rearrangements or loss of heterozygosity.
05.02.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0
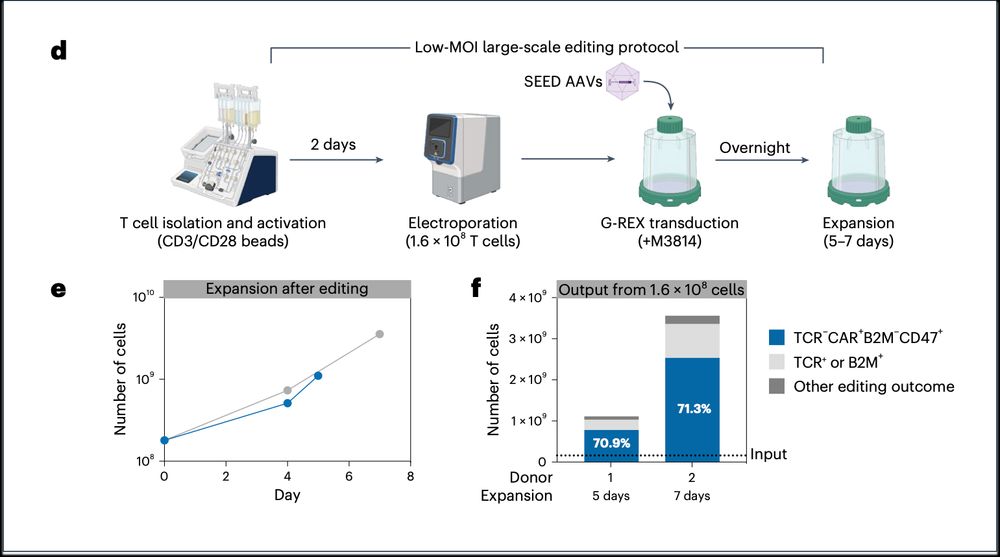
8/ SEED process is highly scalable! We demonstrated that it can be used in clinical-scale manufacturing, generating >2.5 billion fully edited T cells at two loci in just 7 days!
05.02.2025 19:03 — 👍 2 🔁 0 💬 2 📌 0
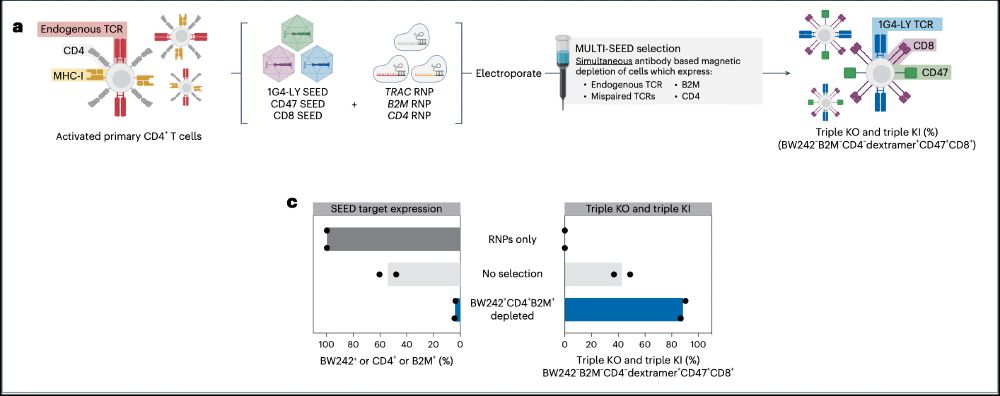
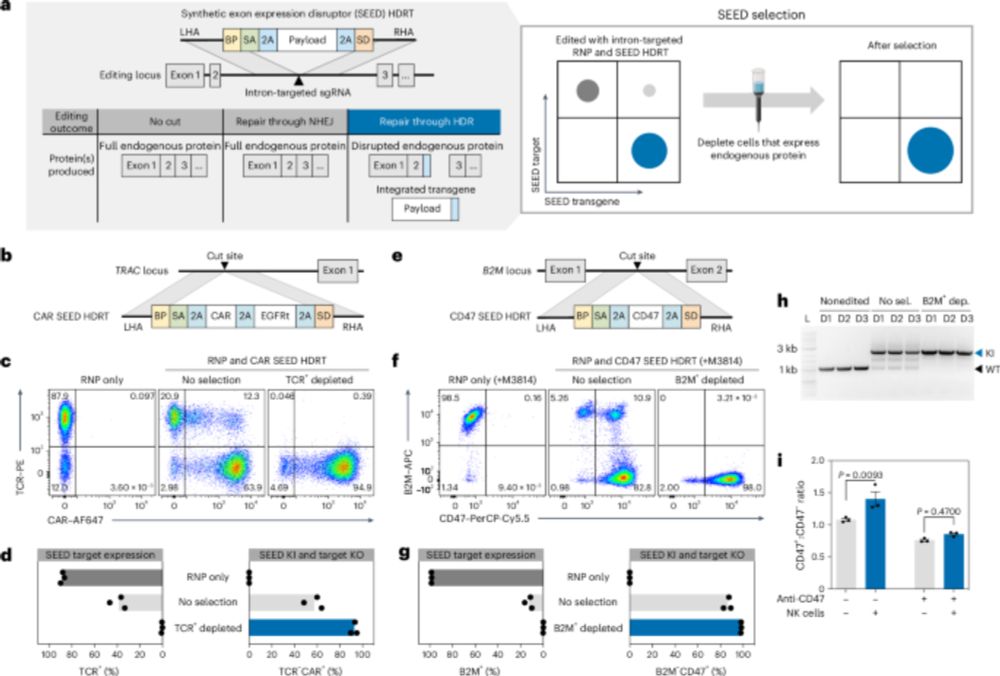
7/ We next wanted to fully reprogram CD4 T cells for TCR specificity, co-receptor swapping and MHC-I with six simultaneous modifications (3 KO & 3 KI). We achieved robust triple KI efficiency (>20%), which was enriched to >90% after SEED.
05.02.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
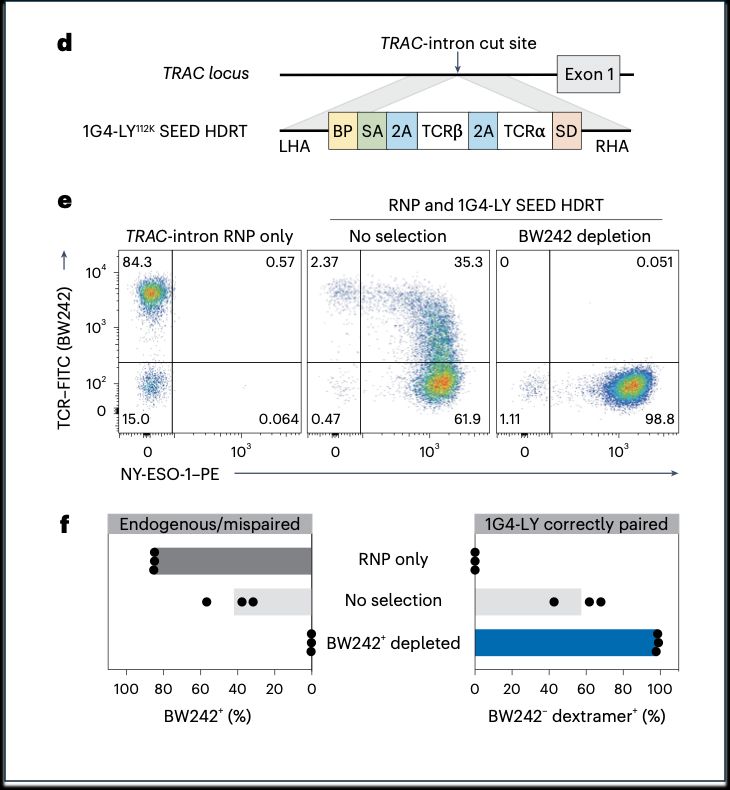
6/ We also tackled TCR mispairing, a major challenge in engineered TCR therapies.
➡️By integrating the epitope-edited TCR, we can identify and eliminate mispaired TCRs and SEED select for cells with proper receptor expression and function!
05.02.2025 19:03 — 👍 0 🔁 0 💬 1 📌 0
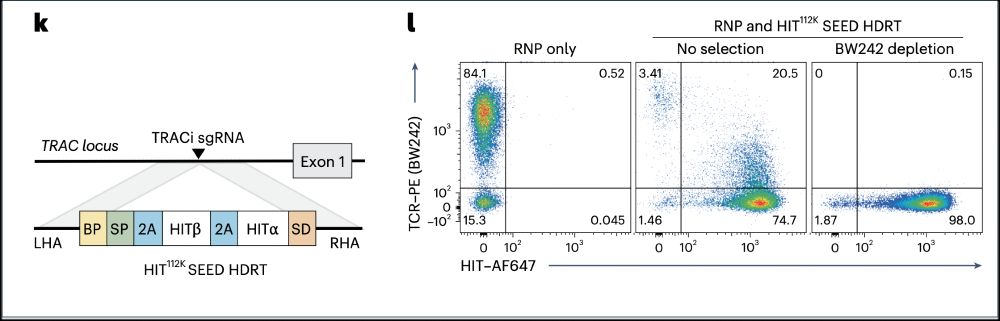
5/ To use SEED on TCR-based receptors (rTCR or HIT), we generated an epitope edited TCR. Using a pool KI screening technology, we identified TCR beta constant chain variants (here D112K) that evade GMP anti-TCR antibodies and enables TCR engineered T cells to be enriched to >95%.
05.02.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
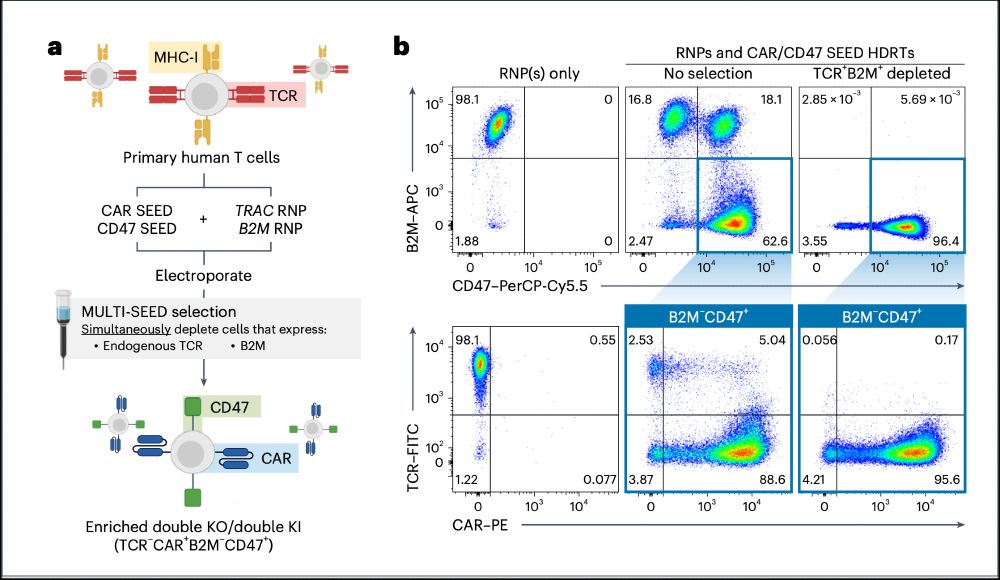
4/ We show that SEED can be multiplexed, enriching for more than 90% purity of double CAR / CD47 KI T cells in a single negative selection step.
05.02.2025 19:03 — 👍 0 🔁 0 💬 1 📌 0
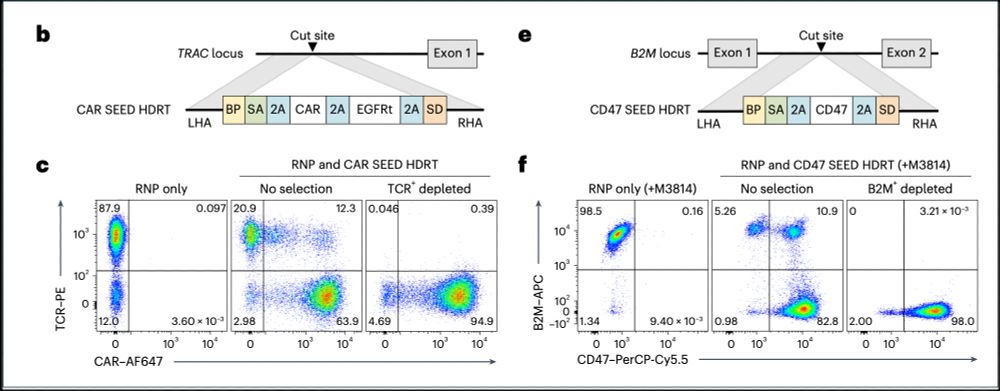
3/ SEED-Selection achieves >95% purity for single modifications. We show we can easily enrich for TRAC-targeted CAR T cells or B2M-targeted CD47 positive T cells through negative selection.
05.02.2025 19:03 — 👍 0 🔁 0 💬 1 📌 0
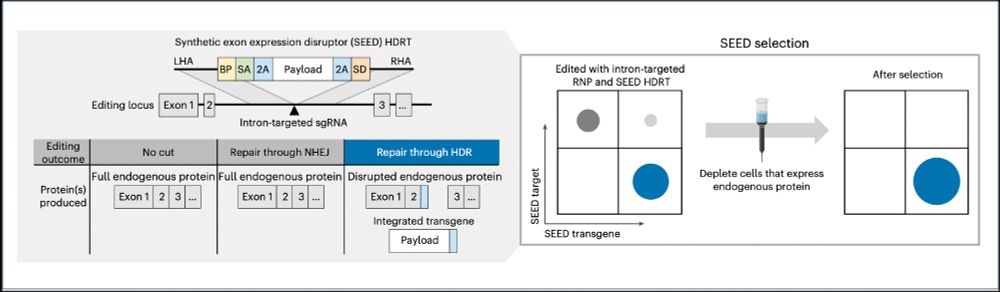
2/ How does it work? We developed a novel class of repair templates called Synthetic Exon Expression Disruptors (SEEDs). SEEDs link successful transgene integration to the disruption of an endogenous surface protein, allowing for easy negative selection.
05.02.2025 19:03 — 👍 0 🔁 0 💬 1 📌 0
1/ T cell engineering is essential for next-gen cell therapies, but multiplexed genome edits result in heterogeneous mixtures of partially edited cells. SEED-Selection allows us to enrich for fully edited T cells through negative selection, leaving engineered cells untouched!
05.02.2025 19:03 — 👍 0 🔁 0 💬 1 📌 0
SEED-Selection enables high-efficiency enrichment of primary T cells edited at multiple loci - Nature Biotechnology
Primary T cells with multiple genetic modifications are rapidly isolated by negative selection.
Excited to share our latest work, now out in
@NatureBiotech
! We introduce SEED-Selection, a powerful method that enables high-efficiency enrichment of T cells edited at multiple loci in a single step 🧵👇
www.nature.com/articles/s41...
05.02.2025 19:03 — 👍 30 🔁 9 💬 2 📌 2
Addgene: pArk313
Plasmid pArk313 from Dr. Justin Eyquem's lab contains the insert Ark313 Rep/Cap and is published in Cell. 2023 Jan 19;186(2):446-460.e19. doi: 10.1016/j.cell.2022.12.022. Epub 2023 Jan 12. This plasm...
10/ Excited to see how this tool will help the field of immunology and gene therapy!
Ark13 on addgene: addgene.org/200257/
📢Read the full paper here: authors.elsevier.com/sd/article/S...
💬Questions? Thoughts? Let’s discuss! ⬇️
04.02.2025 20:50 — 👍 2 🔁 0 💬 0 📌 1
9/ This was a collaboration with the asokan Lab and led by @williamnyberg.bsky.social, Charlotte Wang
and Jon Ark. Huge thanks to our incredible team, collaborators and funders @parkerici.bsky.social @gladstoneinst.bsky.social
@ucsfcancer.bsky.social @pewhealth.bsky.social
04.02.2025 20:50 — 👍 3 🔁 1 💬 1 📌 0
8/ TL;DR—Ark313 enables in vivo T cell engineering:
✅High-efficiency transduction of circulating & tissue-resident T cells
✅Supports CRISPR editing in living mice
✅Enhances cancer immunotherapy via gene KO
✅Enables site-specific transgene integration in T cells
04.02.2025 20:50 — 👍 2 🔁 0 💬 1 📌 0
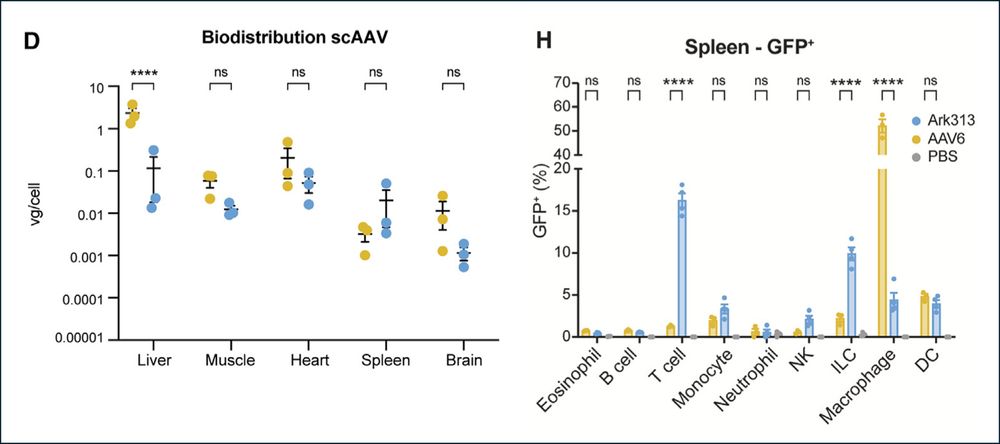
7/ Finally, unlike many AAVs, we found that Ark313 is significantly de-targeted from both the liver and macrophages, reducing off-target effects.
04.02.2025 20:50 — 👍 2 🔁 0 💬 1 📌 0
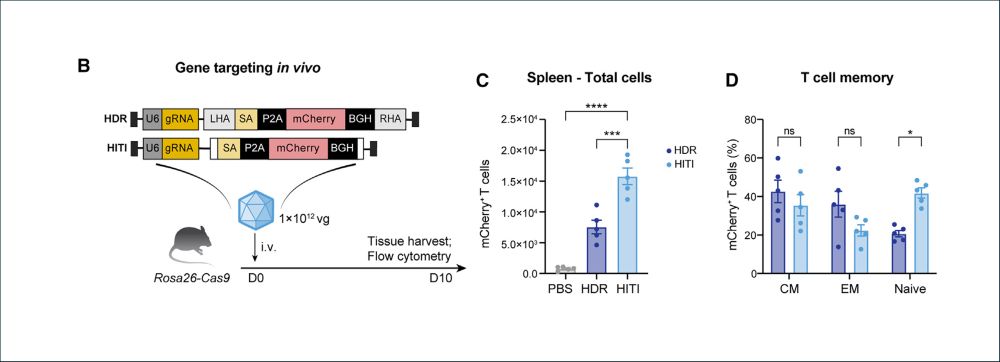
6/ Achieving site-specific integration in vivo is a grail in gene therapy. Ark313 enables targeted integration of large DNA payloads in vivo! Using Homology Directed Repair and Homology-Independent Targeted Integration (HITI), we achieved in vivo KI in a cas9 expressing mouse!
04.02.2025 20:50 — 👍 2 🔁 0 💬 1 📌 0
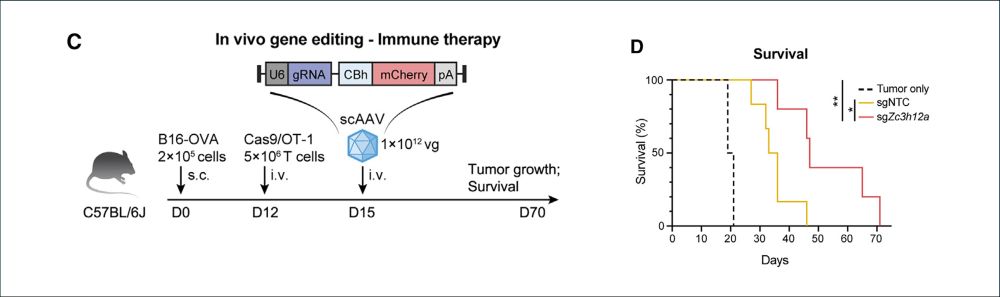
5/ We also tested whether Ark313 could enhance cancer immunotherapy. Knockout of Regnase-1 using Ark313 led to improved T cell function and tumor control in a murine melanoma model.
🔥In vivo gene editing to enhance anti-tumor immunity!
04.02.2025 20:50 — 👍 1 🔁 0 💬 1 📌 0
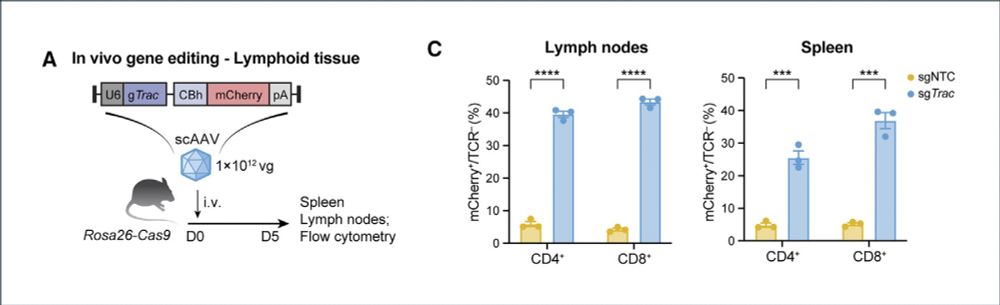
4/ We validated Ark313 for in vivo CRISPR gene editing. A single injection of Ark313 delivering a sgRNA in Cas9-expressing mice resulted in up to 50% KO of the transduced T cells—including tissue-resident cells.
04.02.2025 20:50 — 👍 2 🔁 0 💬 1 📌 0
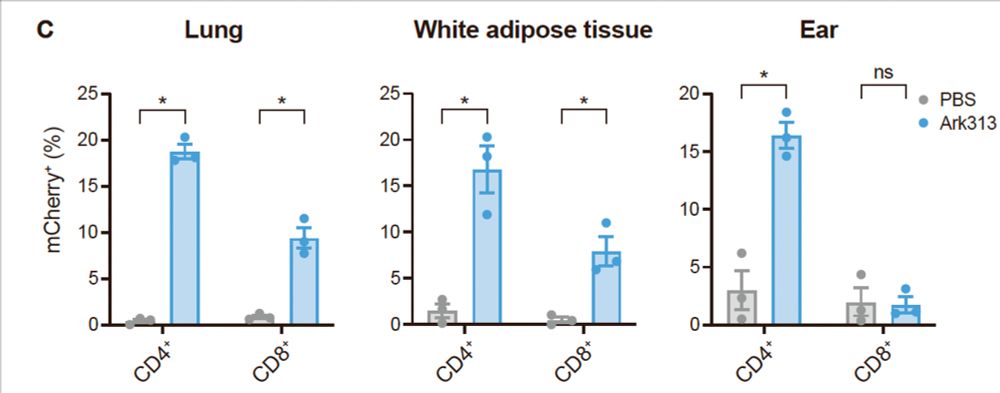
3/ With a single IV injection, Ark313 achieves gene delivery in up to 25% of T cells, in both circulating and tissues, including tissue resident memory! This unlocks new possibilities for studying T cell function in their natural niche. (🙏 to Roberto Ricardo Gonzalez and @arimolofskylab.bsky.social)
04.02.2025 20:50 — 👍 2 🔁 1 💬 1 📌 0
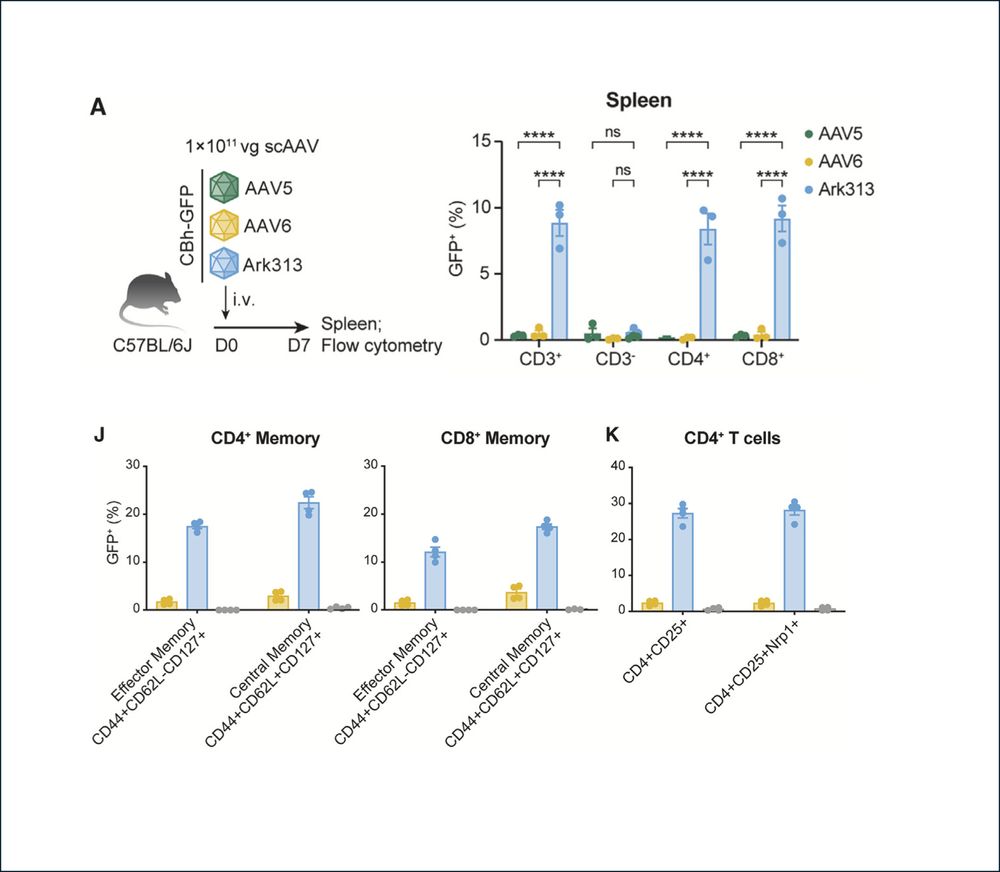
2/ We first show that Ark313 can efficiently transduce T cells in vivo, including naïve, memory and regulatory subsets, outperforming natural AAV serotypes (AAV5 and AAV6).
04.02.2025 20:50 — 👍 1 🔁 0 💬 1 📌 0
1/ T cell engineering is critical for immunology research and therapy, but ex vivo manipulation is complex. We present Ark313, an AAV variant we previously evolved, to directly deliver genes to T cells in vivo—without the need for isolation and re-infusion!
04.02.2025 20:50 — 👍 0 🔁 0 💬 1 📌 0
Wonderful to see the #immunology community growing here.
The first starter pack is full (they cap at 150), so I created a second one. Let me know if you'd like to be added
go.bsky.app/PY5tCas
go.bsky.app/FmERUoD
15.11.2024 12:35 — 👍 196 🔁 99 💬 135 📌 8
Would love to be added! Thanks!!
15.11.2024 21:25 — 👍 1 🔁 0 💬 0 📌 0
Charité Berlin · Baylor College of Medicine Houston · TU Berlin | passionate about genome engineering, CAR T & cell therapies 🧬
Official Bluesky account of the Bendall Lab in the Stanford Department of Pathology and Immunology / Cancer Biology / Stem Cell research and training programs.
Immigrant scientist. Mom of 3 boys. First gen. Immunology and stem cells are cool. Fighting cancer and autoimmunity. She/hers.
Anti-racist feminist, LGTBQIA rights. Climate change is too real.
We’re a clinical-stage programmable cell therapy company focused on realizing solid tumor cell therapies. Our purpose is clear: defeat cancer. Engineering Hope™ through living medicines designed to transform patient outcomes.
Yale Immunobiology PhD student
Neuroimmunology
Associate Editor at Nature Biotechnology. Views are my own.
Transforming routine immune profiling to power the future of medicine. I3H builds on Penn Medicine’s pioneering legacy in immunology to advance discovery across disciplines. Perelman School of Medicine, University of Pennsylvania
🔗 www.med.upenn.edu/i3h
Partnering with scientists to accelerate transformation in cancer research.
News from the Sternberg lab at Columbia University, Howard Hughes Medical Institute.
Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
Visit us at www.sternberglab.org
Translational immunologist using principles of fundamental T cell biology and synthetic biology tools to design T cell therapies -- Assistant Professor @Fred Hutch
Our mission is to accelerate the development of breakthrough immune therapies to turn all cancers into curable diseases.
Neuroscientist. Neuro-Oncologist. Mom.
Macrophage Enthusiast
Assistant Professor @BWH @DFCI
Co-founder TheMyeloidNetwork
Wife and mother x 3
https://guerrierolab.bwh.harvard.edu/
Tumor immunologist, Director of Translational Medicine, Cutaneous Malignancies. Assoc. Professor of Surgery/Microbiology & Immunology at Emory. Fitness is fun @PaulosLab
Cellular Immunologist especially interested in analyzing antigen-specific T cells. Professor at Fred Hutchinson Cancer Center in Seattle.
PhD Student in the NIH-UPenn Immunology Program. Interested in T-cell biology and Immunotherapy!
Immunotherapy R&D - looking to develop immunotherapy, cell and gene therapy, and make an end to sickle cell disease.
Scientific Director of Seattle Children's Brain Tumor Research Program
Neuro-oncologist & Scientist focused on biologic vulnerabilities of #DIPG & #CARTcell #immunotherapy
Assistant Professor @UVA_MIC, @uvaimmunology.bsky.social & #OBGYN; Cancer immunotherapy researcher with a T cell engineering & TME focus; Cancer survivor and research advocate; AACR AMC Past-Chair & SITC ECS member. All views are my own. she/her