Out now! In collaboration with Leifu Chang, we uncover the molecular and structural underpinnings of CRISPR-Cas12f-like RNA-guided transcription systems!
Links to the published articles:
tinyurl.com/55kpavet
tinyurl.com/sk6djwx3
Previous thread for the preprint:
bsky.app/profile/did:...
04.03.2026 20:28 —
👍 60
🔁 25
💬 0
📌 2

Programmable DNA insertion in native gut bacteria
A gene-editing approach enables modification of bacteria within the mouse gut
8/9 Next steps include better delivery, reducing fitness costs, and taking MetaEdit into new ecosystems.
🙏 to all co-authors and to Ivo, @sternberglab.bsky.social, and @harriswang.bsky.social for incredible leadership.
14.11.2025 14:25 —
👍 5
🔁 0
💬 1
📌 0
7/9 MetaEdit opens a promising path for editing individual species inside complex microbiomes.
This approach enables new opportunities to probe microbial functions in vivo, elucidate unculturable taxa, and build microbiome-based therapies.
More to come on the horizon. ⚙️🦠🧬💊
14.11.2025 14:25 —
👍 1
🔁 0
💬 1
📌 0

6/9 MetaEdit also enabled the first genetic modification of SFB, a key host–immune microbe that is famously hard to culture.
By integrating a GFP payload, we visualized edited SFB filaments, a major step toward uncovering how they shape immune development in the small intestine.
14.11.2025 14:25 —
👍 2
🔁 0
💬 1
📌 0

5/9 The result was striking. Edited Bt could now digest inulin with little disruption to the community.
Feeding mice inulin expanded the edited Bt ~30-fold, and removing inulin brought it back down.
A fully reversible, diet-tunable way to control engineered bacteria in vivo.
14.11.2025 14:25 —
👍 2
🔁 0
💬 1
📌 0
4/9 With accurate editing in hand, we asked a simple question:
Can we control engineered gut bacteria using diet?
The answer: yes!
We next used MetaEdit to install a 7.5 kb polysaccharide-utilization locus into the genome of native murine Bt in vivo.
14.11.2025 14:25 —
👍 1
🔁 0
💬 1
📌 0

3/9 In the complex mouse microbiome, MetaEdit accurately edits Bacteroides species (Bt) with 99.8% specificity across gigabases.
This let us recover engineered strains straight from stool by integrating a selective payload & using only a tiny 32-base target sequence as the tag.
14.11.2025 14:25 —
👍 3
🔁 0
💬 1
📌 0
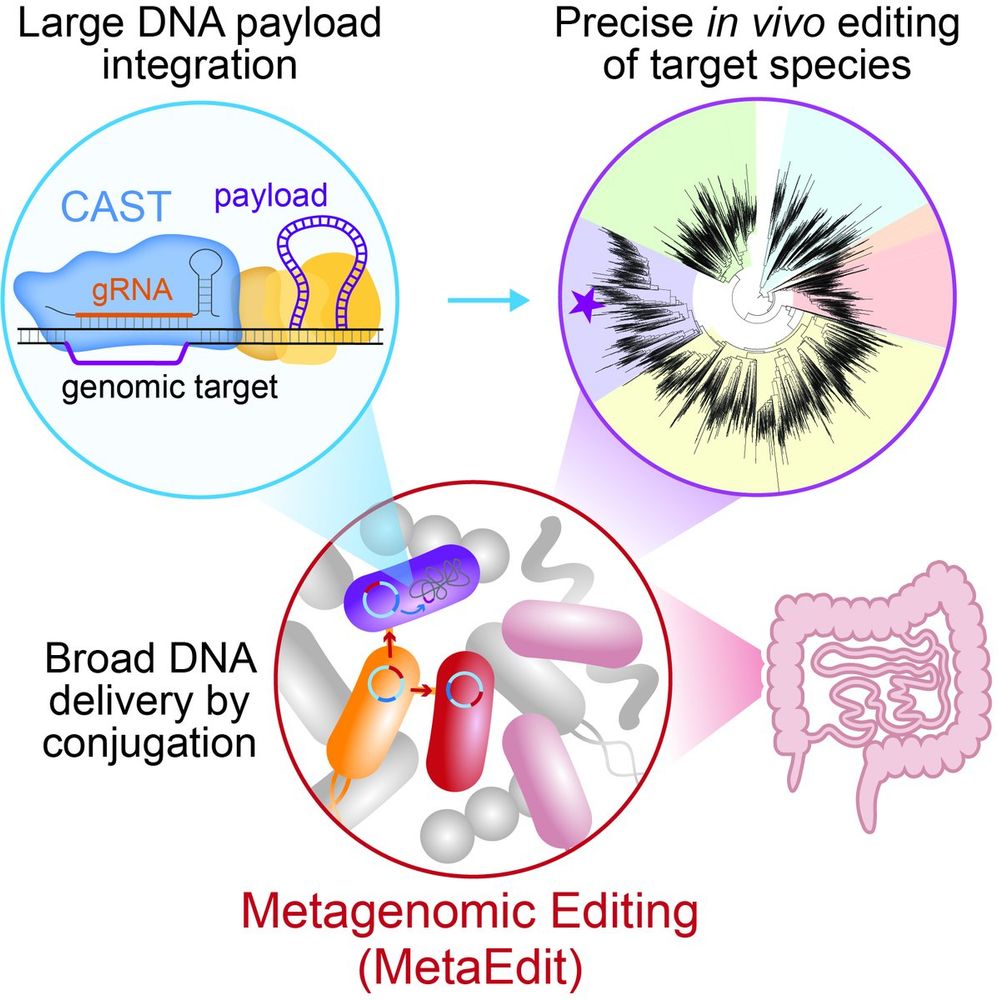
2/9 MetaEdit works by delivering mobile plasmids from a donor E. coli into diverse gut bacteria.
These plasmids encode CRISPR-associated transposases (CAST) to direct RNA-guided insertion of DNA payloads at programmable sites within Gram- and Gram+ members of the gut community.
14.11.2025 14:25 —
👍 4
🔁 0
💬 1
📌 0

Please RT. Post-doc opportunity alert! 💥 closing 10th December.. Come join our team (www.thelowlab.org) at Imperial, London, working on the structure and mechanism of bacterial secretion systems.
For more details and to apply please see
www.imperial.ac.uk/jobs/search-...
04.11.2025 17:19 —
👍 36
🔁 39
💬 2
📌 1
10/10 Many thanks to all co-authors for their collective efforts in bringing this story to fruition! And a special thanks to @shsternberg.bsky.social for continually fostering a spirit of curiosity, creativity, and rigor in all aspects of our work.
17.10.2025 17:22 —
👍 14
🔁 0
💬 1
📌 0
9/10 Collectively, our findings suggest that telomerase-like activity emerged in an ancient bacterial ancestor, and was co-opted in early organisms with linear genomes to set the stage for the evolution of modern eukaryotes.
17.10.2025 17:22 —
👍 14
🔁 3
💬 1
📌 0

8/10 Furthermore, when equipped with the template sequence from the telomerase RNA, DRT10 readily synthesized telomeric DNA repeats.
17.10.2025 17:22 —
👍 13
🔁 3
💬 1
📌 0

7/10 We teamed up with RT aficionado @pentamorfico.bsky.social from the Rafa Pinilla-Redondo lab to build a new phylogenetic tree of RTs across all domains of life.
Remarkably, this revealed that the DRT enzymes are bacterial homologs of TERT.
17.10.2025 17:22 —
👍 16
🔁 2
💬 2
📌 1

6/10 We found that DRT10 synthesizes tandem-repeat cDNAs through a mechanism strikingly reminiscent of DNA repeat addition by telomerase. But does this similarity represent convergent evolution or shared ancestry?
17.10.2025 17:22 —
👍 6
🔁 4
💬 1
📌 0

De novo gene synthesis by an antiviral reverse transcriptase
Defense-associated reverse transcriptase (DRT) systems perform DNA synthesis to protect bacteria against viral infection, but the identities and functions of their DNA products remain largely unknown....
5/10 Following our recent work on the DRT2 and DRT9 antiviral immune systems in bacteria (linked below), which revealed intricate mechanisms of RNA-templated repetitive DNA synthesis, we began studying a new system, DRT10.
DRT2: science.org/doi/10.1126/...
DRT9: nature.com/articles/s41...
17.10.2025 17:22 —
👍 6
🔁 0
💬 1
📌 0
4/10 Telomerase is found in nearly all eukaryotes and acts as a critical safeguard against genome instability from progressive DNA loss. Its aberrant activation is also key to the growth of cancer cells.
And yet, the evolutionary origin of telomerase has long remained unresolved.
17.10.2025 17:22 —
👍 6
🔁 0
💬 1
📌 0

3/10 Forty years ago, Carol Greider and Elizabeth Blackburn discovered an enzyme that solves this problem. Telomerase, which comprises a reverse transcriptase (TERT) and RNA (TR), directly extends chromosome ends by adding DNA repeats templated by the TR.
17.10.2025 17:22 —
👍 9
🔁 1
💬 1
📌 0
2/10 Linear chromosomes shorten with each round of cell division. Famously known as the “end-replication problem,” this phenomenon eventually leads to cellular senescence and is one of the hallmarks of aging.
17.10.2025 17:22 —
👍 6
🔁 0
💬 1
📌 0
16/16 This work was a big team effort and fun collaboration! Thanks to everyone: Matt, Egill, Tanner, Jing, Zaofeng, Americo, Florian, Hamna, Ryan, Nick, Israel and our stellar mentor Sam for making this project possible!
24.07.2025 19:58 —
👍 0
🔁 0
💬 0
📌 0
15/16 In summary, we shed light on the biological role of a naturally occurring version of CRISPR interference (CRISPRi), that emerged in gut-associated bacteria to benefit multiple aspects of their lifestyle.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
14/16 The repeated exaptation of TnpB to regulate flagellin suggests strong evolutionary pressure favoring RNA-guided control of flagellin expression, particularly in gut-associated bacterial species.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
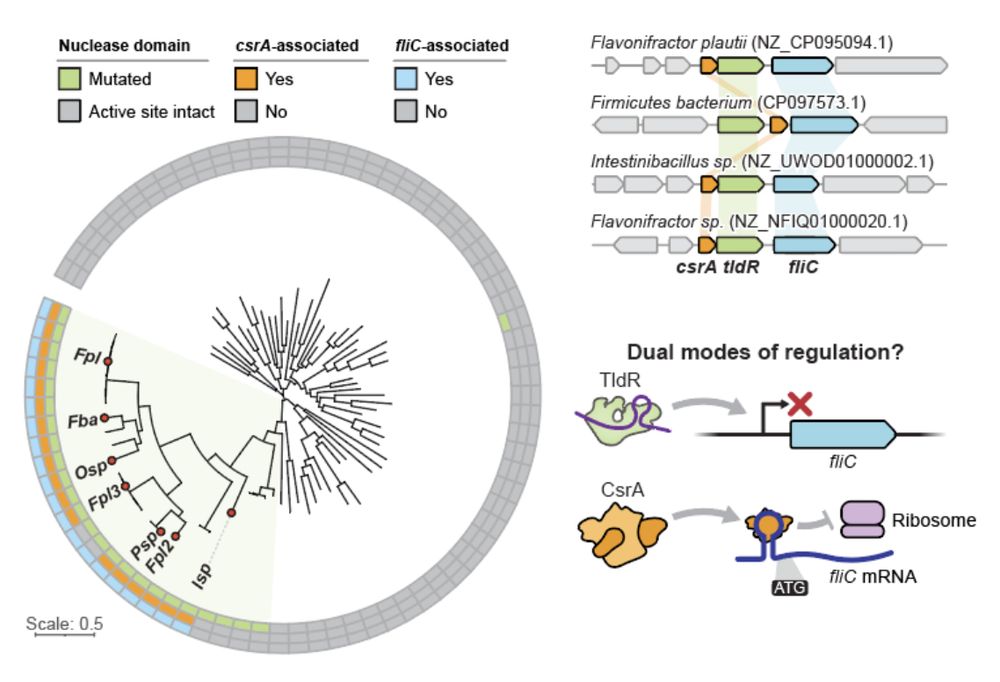
13/16 In other key members of the human gut microbiome, flagellin-regulating TldRs can be found additionally associated with the translational repressor CsrA. The coordinated action of TldR and CsrA suggests a dual mode of both transcriptional and post-transcriptional gene regulation.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
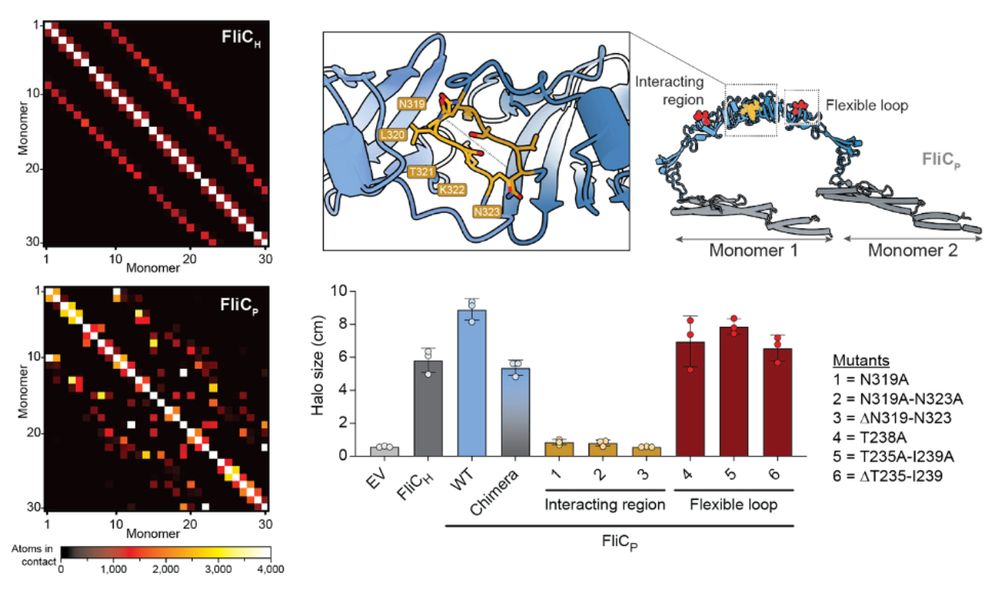
12/16 The extra domain stabilizes the filament by increasing inter-subunit contacts. Structure-guided mutagenesis confirmed that this domain is essential to improve motility of the lysogen. We believe it could similarly explain the decreased activation of TLR5.
24.07.2025 19:58 —
👍 1
🔁 0
💬 1
📌 0
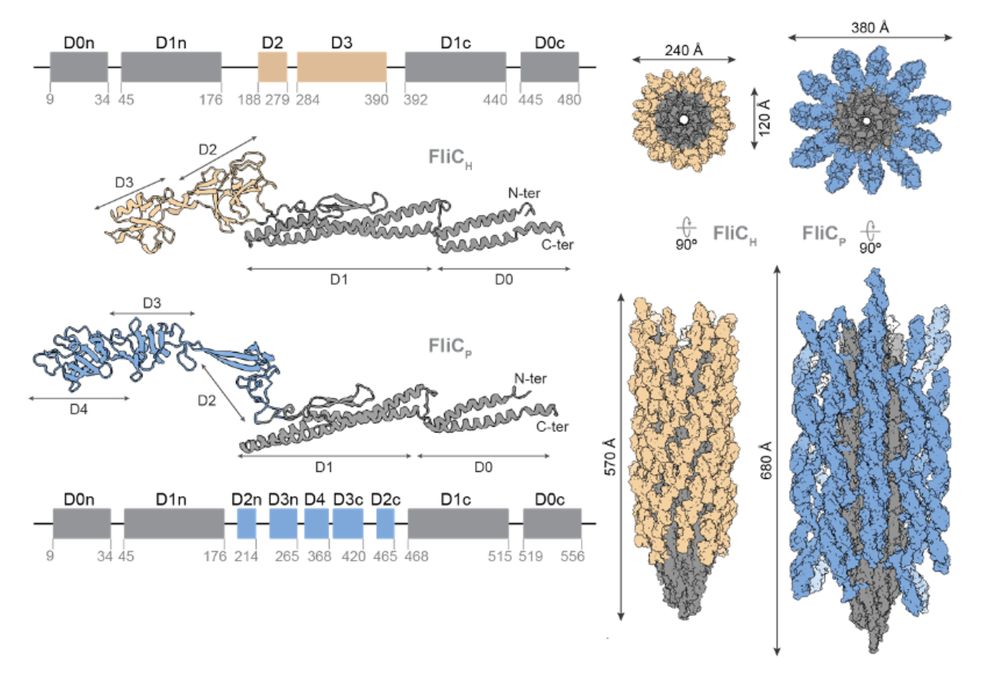
11/16 We found that both flagellar filaments have a drastically different structure. The prophage flagellar filament is significantly thicker and presents a surprising mesh-like structure. This is due to the presence of an extra domain in the prophage flagellin.
24.07.2025 19:58 —
👍 1
🔁 0
💬 1
📌 0
10/16 To try to link these phenotypic observations, we teamed up once again with @piratefernandez.bsky.social and decided to inspect the structures of the prophage flagellin in comparison to their host counterpart.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
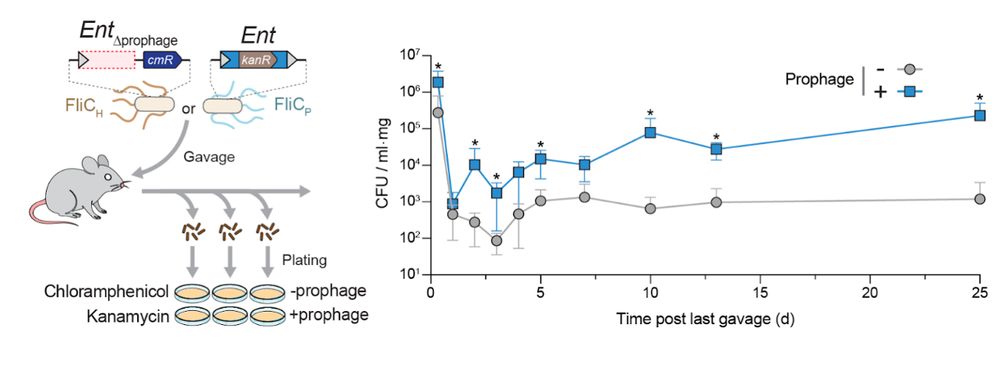
9/16 The phenotypic effects of lysogenization result in a consequential ecological outcome: enhanced colonization of the murine gut, illustrating the advantage of flagellar remodeling in a host-associated environment.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0
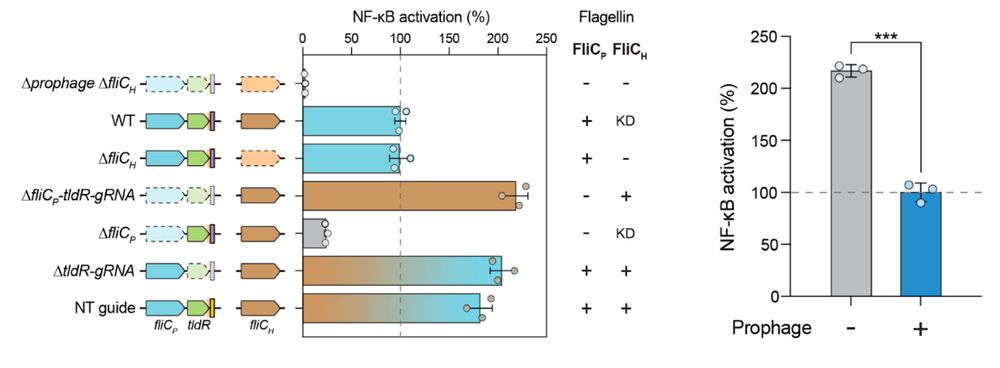
8/16 We also find that lysogenic conversion by FRφ reduces activation of TLR5, a receptor that detects flagellin to initiate the immune response, suggesting that lysogenization may facilitate immune evasion.
24.07.2025 19:58 —
👍 0
🔁 0
💬 1
📌 0

















