
🚨📢📄 Article in press in Genome Biology doi.org/10.1186/s130...
We introduce panREPET, a reference-free pipeline to detect shared transposable element (TE) insertions across pangenomes and retrace their evolutionary dynamics #TEsky 🧵👇
@p-bourguet.bsky.social
Biologist studying transposons, gene regulation and chromatin in the Jullien group. Postdoc @ IBMP (Strasbourg, France). @ibmp-cnrs.bsky.social

🚨📢📄 Article in press in Genome Biology doi.org/10.1186/s130...
We introduce panREPET, a reference-free pipeline to detect shared transposable element (TE) insertions across pangenomes and retrace their evolutionary dynamics #TEsky 🧵👇

How could a simple self-replicating system emerge at the origins of life? RNA polymerase ribozymes can replicate RNA, but existing ones are so large that their self-replication seems impossible. Could they be smaller?
Excited to share our latest work in @science.org on a new small polymerase.
1/n
Check out this story by Handler, Brennecke et al. on how the piRNA pathways distinguish transposable element transcripts. Beautiful work!
13.02.2026 15:46 — 👍 10 🔁 1 💬 0 📌 0Thrilled to announce I've been awarded a Marie Skłodowska-Curie Postdoctoral Fellowship! 🇪🇺 I'll be joining @pejullien.bsky.social's group at @ibmp-cnrs.bsky.social in Strasbourg. Excited for what's ahead! 🎉
11.02.2026 12:38 — 👍 25 🔁 1 💬 6 📌 0This week in @science.org, a celebration of plant pangenomes. A pangenome analysis for massively polyploid sugarcane species, and one for Brassica rapa giving insight into subspeciation.
What's all the fuss about pangenomes? Pamela and Douglas Soltis explore this in an insightful Perspective(1/4)

'The Centre for Research in Cellular Biology of Montpellier (CRBM) is recruiting Junior/Senior Group Leaders to shape its research in the coming decade.
The deadline for application is 31st of March 2026.'
#sciencejobs
focalplane.biologists.com/cell-biology...

Here we go again! Join us in Vienna, May 21–22, for two days of plant science—talks, discussions, and celebration of what makes plants both beautiful and essential 💚Full program + registration here: www.oeaw.ac.at/gmi/news-eve...
Come be part of it — each of you counts!
I am excited to offer a postdoctoral research position in my lab in Oxford studying the distribution of small RNAs during plant sexual reproduction. shorturl.at/7Goxt
Reposts appreciated!
Obligatory cat video to brighten your day. :-)

📢 Three new #bioRxiv preprints from our team on holocentric chromosomes.
Together, they connect centromere repeat evolution, karyotype dynamics, and meiotic recombination outcomes, revealing how holocentric genomes evolve and function. 🧬👇

Dominant contribution of Asgard archaea to eukaryogenesis.
tldr; Best guess, ~ 50% of conserved eukaryotic protein families are from from Asgard archaea!
#science #biology #evolution #nature #bioinformatics
www.nature.com/articles/s41...


After a short break in our webinars we are delighted to be back up & running this month!
First up, on Jan 30th we will be joined by @lucas.farnunglab.com from @harvardmed.bsky.social 🔬
Shortly after, on Feb 13th we will be hosting @heardlab.bsky.social from @crick.ac.uk 🧬
Registration details 👇

The (Yoav) Voichek lab has opened its gates at the Weizmann Institute, and is actively recruiting students and researchers at all levels - come explore gene regulation and computational genomics in a fun, friendly sprouting lab 🤗🥼⚗️🧪
www.weizmann.ac.il/plants/voichek

New Year, New Paper!🎊
Pervasive cis-regulatory co-option of a transposable element family reinforces cell identity across the mouse immune system
www.biorxiv.org/content/10.6...
Centerpiece of Jason Chobirko's PhD, talented PhD student co-mentored by Andrew Grimson & me. Really excited about it!🧵

Have we been overstating the role of transposable elements in adaptation to local or rapidly changing environmental conditions? Happy to share my (somewhat unpopular?) opinion paper on this matter: www.sciencedirect.com/science/arti...
Thanks to those who already shared the link!
REFEREE 1: “I conclude with a simple, direct statement - this is the best paper I have read all year!”
#TransparentPeerReview
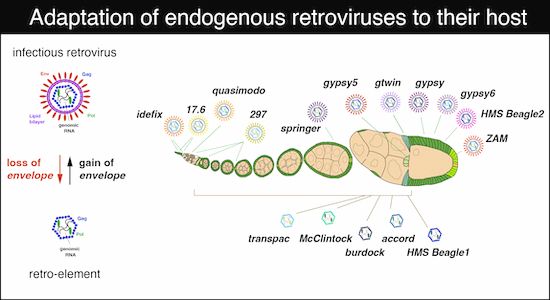
Co-evolving infectivity and expression patterns drive the diversification of endogenous retroviruses
@juliusbrennecke.bsky.social et al
www.embopress.org/doi/full/10....
Intrigued by a long-standing conundrum in small RNA biology—how nuclear Argonaute proteins silence transposons when they *need* target transcription for their own recruitment—we studied the piRNA pathway.
And found a hidden RNA-decay axis from Piwi to the RNA exosome.

Applications for our international #PhD program open until 11th Jan. Come work with us @uoe-igc.bsky.social and @hannahlong.bsky.social to understand DNMT3B in ICF syndrome or with our wonderful colleagues.
Details here:
institute-genetics-cancer.ed.ac.uk/igc-graduate...
#epigenetics Please repost 🙏

While stories of singular DNA changes that drove evolution of human brain/behaviour remain seductive, advances across multiple fields of biology cast doubt on such simplistic narratives of our origins. A new paper from my lab shows how biobanks may speak to this fundamental question.🧪
Explainer🧵👇1/n
Closing out my year with a journal editor shocker 🧵
Checking new manuscripts today I reviewed a paper attributing 2 papers to me I did not write. A daft thing for an author to do of course. But intrigued I web searched up one of the titles and that's when it got real weird...

Position du lever du soleil en fonction des saisons
Aujourd'hui c'est le solstice d'hiver dans l'hémisphère Nord. Les journées vont commencer à rallonger! Le soleil ne se lève à l'Est (centre de l'image avec Nord à gauche, Sud a droite) que lors des équinoxes. La position du lever change un peu tous les jours! (©Z. Al-Abbadi)
21.12.2025 08:48 — 👍 969 🔁 260 💬 16 📌 11
1/ I’ve reviewed hundreds of bioinformatics GitHub repos in my career.
Here’s the brutal truth: most tool documentation fails the people it’s meant to help.
And it’s not because the algorithms are bad.

Have you ever wondered how new DNA methylation patterns are established?
Paradigm shift ahead! We discovered a new mode of DNA methylation targeting in plants that relies on transcription factors and sequence motifs rather than chromatin modifications to regulate the methylome. rdcu.be/eQ6L5
1/8

An early Christmas present for those interested in chromatin and transcription! Fantastic work from @au-ho-yu.bsky.social and @aleksszczurek.bsky.social . Thanks to Inge and Michiel for their help. Please repost!
www.biorxiv.org/content/10.6...

Transposable elements can insert into genes, disrupting protein-coding potential. Researchers discover a mode of RNA processing, ‘SOS splicing’, that provides a quick fix @nature.com @harvardmed.bsky.social
www.nature.com/articles/d41...
www.nature.com/articles/s41...
Great news, congratulations!
09.12.2025 17:14 — 👍 1 🔁 0 💬 1 📌 0
Looking for a postdoc in computational biology? We are looking for a postdoc candidate apply for the fellowship 'Juan de la Cierva' for a project on long-read transcriptomics, multiomics, spatial transcriptomics, single-cell long-reads. Deadline is approaching!! (December 10th)
08.12.2025 12:06 — 👍 4 🔁 7 💬 0 📌 0
🔬 Call to create junior research groups at the Institut Pasteur
Focus: Infectious diseases, host-microbe interactions, vaccines
Special interest: AI methodologies
📅 Deadline: Feb 9, 2026
👥 2-12 years post-PhD
Apply now 📝 research.pasteur.fr/en/call/crea...
#JobOpportunity #Research

Congratulations the first story from your lab Marco! To all co-authors and to Gesa for the stunning pictures 😍
02.12.2025 15:42 — 👍 3 🔁 0 💬 1 📌 0
Preprint alert! It is my great pleasure to announce the first manuscript from the lab, a story that started @gmivienna.bsky.social and was mainly accomplished by the intrepid @gesahoffmann.bsky.social at @mpi-mp-potsdam.bsky.social. A brief thread with our findings
www.biorxiv.org/content/10.1...
Have you ever wondered how it is possible that systemically infected plants produce (a certain percentage of) healthy progeny? 💚
Check out the first preprint of the @incavirus.bsky.social lab and first preprint of my postdoc! 🍀
We'd be very happy about feedback and discussions!