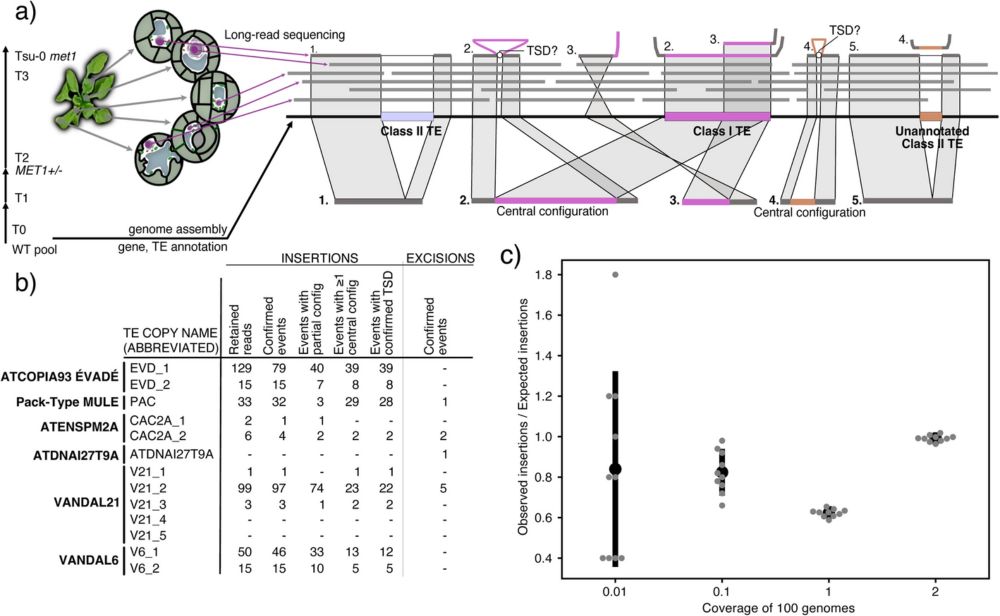
An early Christmas present from our team for all of the tomato 🍅 & recombination fans:
Chromosome-scale Solanum pennellii and Solanum cheesmaniae genome assemblies reveal structural variants, repeat content and recombination barriers of the tomato clade
biorxiv.org/content/10.648…
21.12.2025 15:00 —
👍 27
🔁 11
💬 2
📌 0

📣Plant Meiosis Meeting is back for its second edition‼️
Save the date: 9–11 September 2026, Poznań, Poland🌱
More details coming soon🤩
06.12.2025 20:42 —
👍 18
🔁 10
💬 0
📌 1
How do plants control the genes needed for successful sexual reproduction? Our new work uncovers the DREAM complex as a crucial regulator of meiotic recombination in Arabidopsis. Without DREAM, plants are sterile—key recombination genes like MER3 are switched off...
05.11.2025 15:28 —
👍 11
🔁 7
💬 1
📌 0
🌿🔍Paper alert‼️
A fantastic study from James Higgins’ group on SCEP3 – a newly identified component of the synaptonemal complex in plants!
Happy that our team could be part of this! 😄🧬
19.11.2025 20:49 —
👍 3
🔁 0
💬 0
📌 0
⏰ Last week to apply!
Don’t miss the chance to join our lab and explore the fascinating world of meiotic recombination 🌿
👉 Deadline: Nov 1, 2025
25.10.2025 05:48 —
👍 6
🔁 6
💬 0
📌 0

Meiotic cohesin RAD21L shapes 3D genome structure and transcription in the male germline
Deletion of the meiotic subunit RAD21L alters 3D chromatin structure and transcription in the mammalian male germ line.
🚨 New from our lab in #ScienceAdvances!
RAD21L cohesin acts as both a 3D genome architect and a transcriptional gatekeeper in the male germline. Its loss disrupts both structure and function—highlighting the tight link between genome folding and gene regulation
www.science.org/doi/10.1126/...
02.10.2025 01:35 —
👍 43
🔁 11
💬 5
📌 2
Sure!!! Meiosis rules!
Fingers crossed for your paper 🤞
01.10.2025 19:27 —
👍 2
🔁 0
💬 0
📌 0
A great story from @chloegirard.bsky.social, confirming and extending our findings on how interhomolog polymorphism shapes crossover distribution in plants 🌱
01.10.2025 17:03 —
👍 4
🔁 1
💬 1
📌 0

🚀 We’re hiring a Postdoc!
Join our group in Poznan, Poland to study meiotic crossover recombination in plants 🌱 Highly motivated & enthusiastic candidates are welcome!
📅 Deadline: Nov 1, 2025
🔗 ibmib.web.amu.edu.pl/wp-content/uploads/2025/09/Postdoc_position-2025-Ziolkowskis-Lab.pdf
27.09.2025 05:03 —
👍 23
🔁 23
💬 0
📌 1
Thrilled to see our paper featured on the cover!!! 🎉🌽
20.09.2025 11:15 —
👍 19
🔁 2
💬 0
📌 0
Thank you so much, Ian! 🙏 I’m really happy I had the chance to work with you to discover this phenomenon ✨, and now to confirm it in a crop as well! 🌽🎉
20.09.2025 11:05 —
👍 2
🔁 0
💬 0
📌 0
Thank you so much!
13.09.2025 09:23 —
👍 1
🔁 0
💬 0
📌 0

Hybridization and introgression are major evolutionary processes. Since the 1940s, the prevailing view has been that they shape plants far more than animals. In our new study (www.science.org/doi/10.1126/...
), we find the opposite: animals exchange genes more, and for longer, than plants
12.09.2025 07:54 —
👍 203
🔁 121
💬 3
📌 3
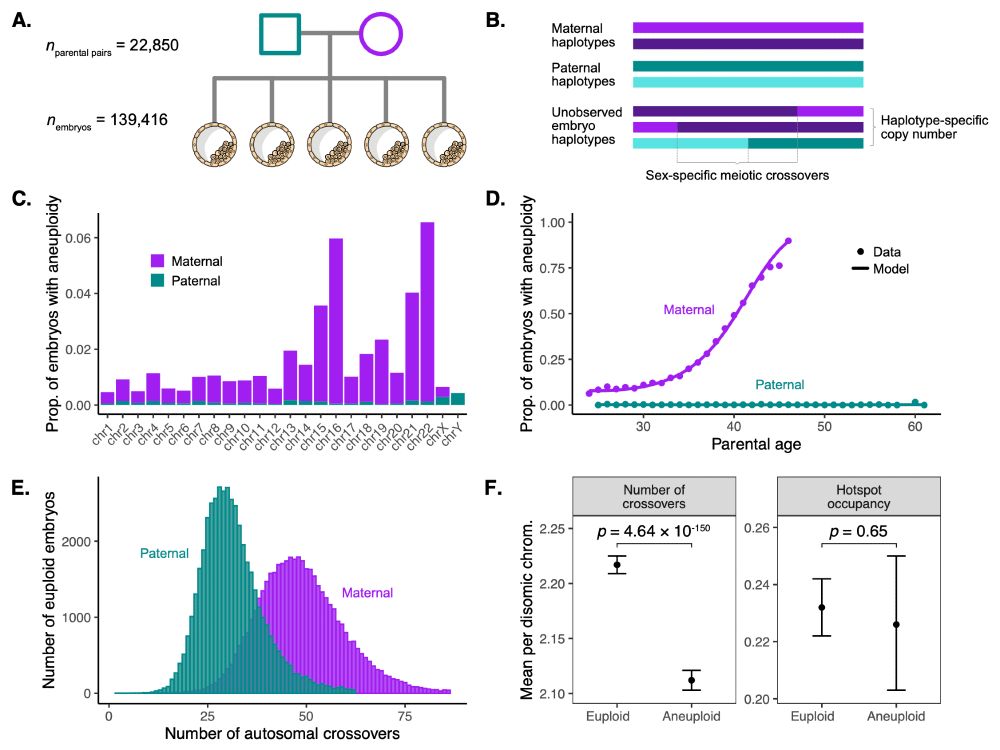
Aneuploidy is the leading cause of pregnancy loss. In work led by @saracarioscia.bsky.social and @aabiddanda.bsky.social, we reanalyzed genetic testing data from 139,416 IVF embryos to discover variants associated with recombination phenotypes and aneuploidy risk. www.medrxiv.org/content/10.1...
07.04.2025 14:00 —
👍 89
🔁 38
💬 1
📌 3
Cryogenic electron microscopy has been used to determine the detailed structure of an intermediate state called a “D-loop” that forms when strands of DNA are exchanged during homologous recombination.
buff.ly/pZZfIso
10.09.2025 22:28 —
👍 17
🔁 5
💬 0
📌 0
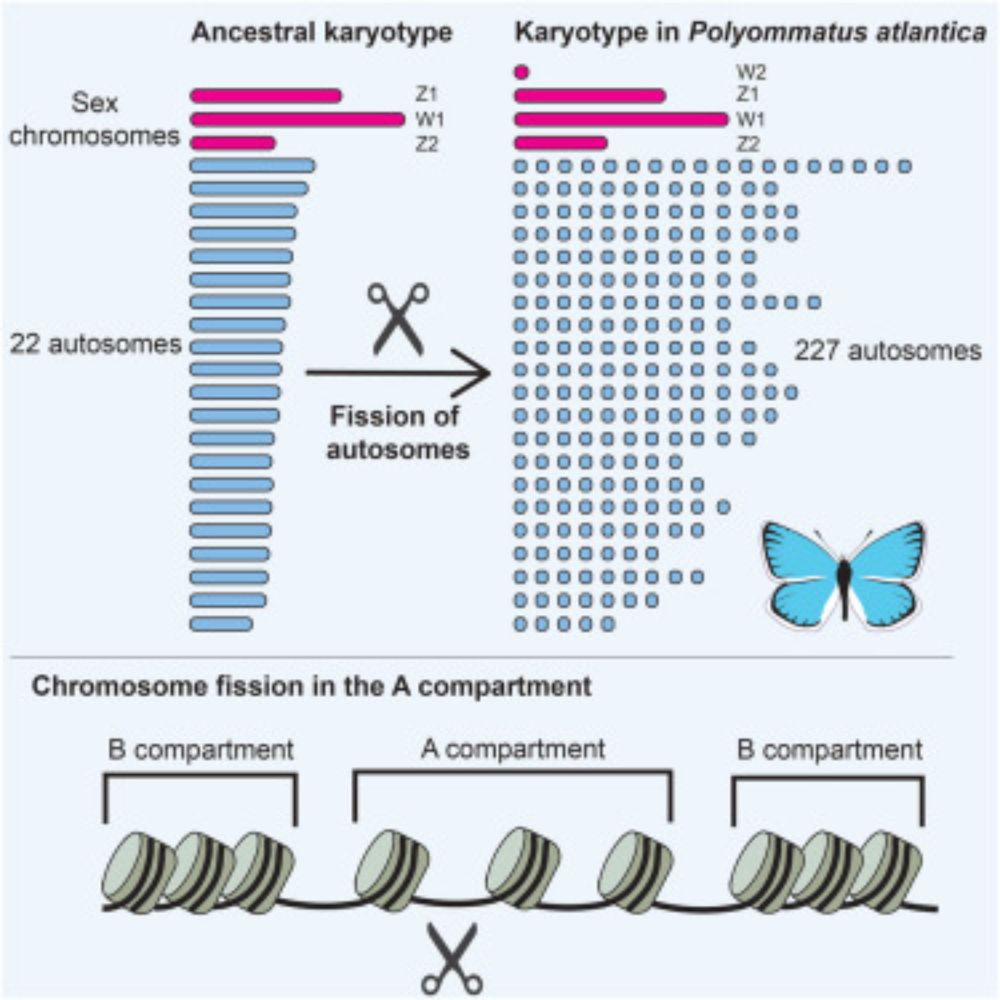
Constraints on chromosome evolution revealed by the 229 chromosome pairs of the Atlas blue butterfly
The genome of the Atlas blue butterfly contains ten times more chromosomes than most
butterflies, and more than any other known diploid animal. Wright et al. show that
this extraordinary karyotype is ...
How many chromosomes can an animal have?
In our paper out now in @currentbiology.bsky.social we show that the Atlas blue butterfly has 229 chromosome pairs- the highest in diploid Metazoa! These arose by rapid autosome fragmentation while sex chromosomes stayed intact.
www.cell.com/current-biol...
11.09.2025 15:21 —
👍 214
🔁 99
💬 4
📌 6
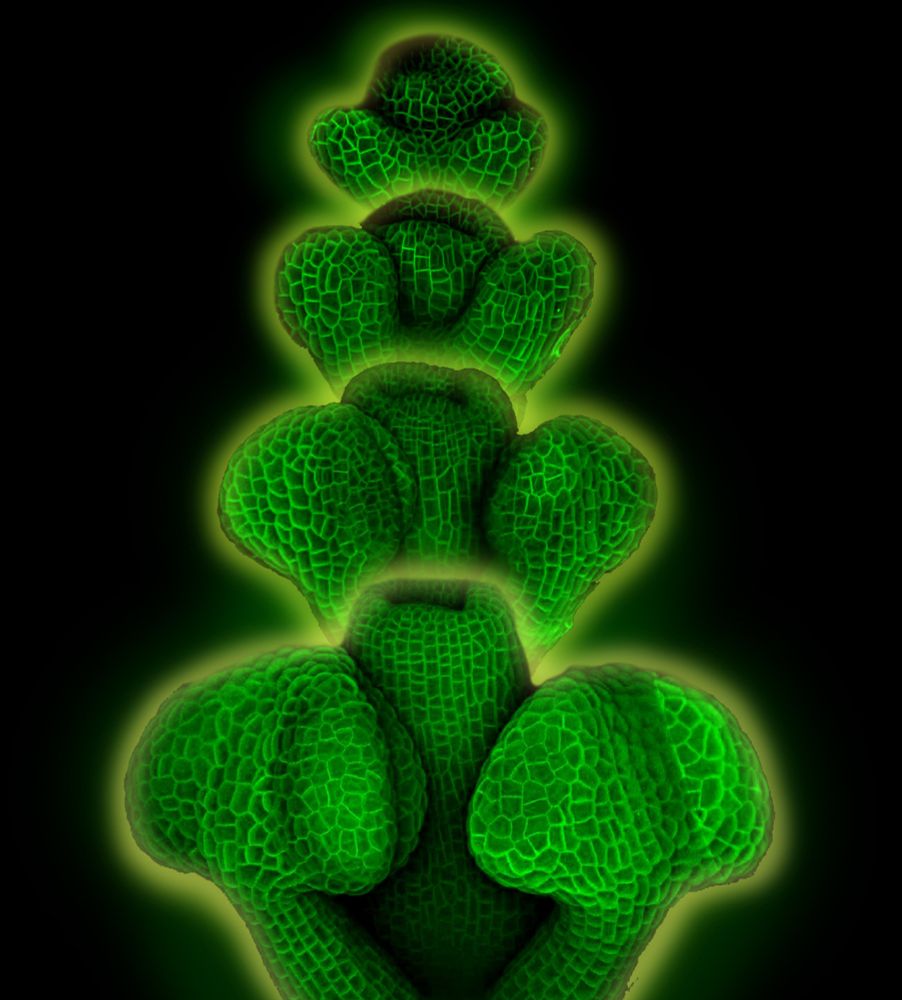
Do you want to know how to live-image internal floral organs with confocal microscope for up to 2 weeks?
Check out our protocol:
bio-protocol.org/en/bpdetail?...
#Arabidopsis #Flower
18.12.2024 13:24 —
👍 129
🔁 41
💬 2
📌 3
Interesting approach!
07.09.2025 11:52 —
👍 0
🔁 0
💬 0
📌 0
Thrilled to share our latest work on meiotic recombination, where we mapped rates and distributions by sequencing thousands of individual sperm. This study was led by Stevan Novakovic and @caitlinharris.bsky.social , in collaboration with @davisjmcc.bsky.social and Cynthia Liu.
05.09.2025 03:49 —
👍 16
🔁 10
💬 2
📌 0
Thanks Wayne!🙏
05.09.2025 09:05 —
👍 0
🔁 0
💬 0
📌 0
Thrilled to see such a thoughtful commentary on our paper by Maria Cuacos & Stefan Heckmann 🙌 Many thanks! 🔬🙏
04.09.2025 10:29 —
👍 10
🔁 2
💬 1
📌 0
This work was made possible through a great collaboration with Mikhail Mikhailov from Moldova 🇲🇩 and the fantastic efforts of all co-authors – special thanks to @franz_boideau, @SzymanskaLejman and Vasile Botnari 👏🌱
02.09.2025 10:19 —
👍 3
🔁 0
💬 0
📌 0