
When plotting, LLMs see what they expect to see - Posit
Data science agents need to accurately read plots even when the content contradicts their expectations. Our testing shows today's LLMs still struggle here.
Introducing bluffbench, a new tool to evaluate how well LLMs actually see data plots.
When we trick LLMs with secret #RStats transformations, they can miss the visual contradiction.
bluffbench helps us measure this "blind spot" in AI coding agents. Learn more: posit.co/blog/introdu...
19.11.2025 16:55 — 👍 22 🔁 11 💬 1 📌 2

Opening of #PMS2025 by @vcarryon.bsky.social @josraaijmakers.bsky.social @microsos.bsky.social
03.11.2025 17:41 — 👍 7 🔁 1 💬 0 📌 0
Check out the quality of segmentation by #OneRosette, even when leaves are dying to DC3000. Come and see nuts and bolts of this deep learning prompt-engineering by PhD candidate Felicià Maviane @lipme-toulouse.bsky.social on the 24/10/2025 13h #Margulis
15.10.2025 08:53 — 👍 24 🔁 8 💬 0 📌 0
YouTube video by VPRO Tegenlicht
Wat DNA uit slootwater ons vertelt over jouw onbekende voorouders | VPRO Tegenlicht
Hoe is al het complexe leven op aarde eigenlijk ontstaan?
09.10.2025 07:49 — 👍 6 🔁 2 💬 0 📌 2
Accurate MAG reconstruction from complex soil microbiome through combined short- and HiFi long-reads metagenomics www.biorxiv.org/content/10.1... #jcampubs
15.09.2025 15:51 — 👍 6 🔁 2 💬 0 📌 0
CompareM2 is a genomes-to-report pipeline for comparing microbial genomes academic.oup.com/bioinformati... #jcampubs
15.09.2025 16:27 — 👍 3 🔁 3 💬 0 📌 0

ggplot2 4.0.0
A new major version of ggplot2 has been released on CRAN. Find out what is new here.
I am beyond excited to announce that ggplot2 4.0.0 has just landed on CRAN.
It's not every day we have a new major #ggplot2 release but it is a fitting 18 year birthday present for the package.
Get an overview of the release in this blog post and be on the lookout for more in-depth posts #rstats
11.09.2025 11:20 — 👍 850 🔁 281 💬 9 📌 51
Sylph-tax - Documentation for sylph - ultrafast, precise metagenomic profiling
New sylph pre-built databases + taxonomy available for:
- GTDB-R226 (143k prok. species)
- GlobDB-R226 (>300k prok. species, thanks @daanspeth.bsky.social )
- UHGV (Unified Human Gut Virome Catalog, thanks @apcamargo.bsky.social )
Must update sylph-tax; see docs (sylph-docs.github.io/sylph-tax/)
23.07.2025 14:11 — 👍 38 🔁 16 💬 1 📌 1
New preprint from the lab: "Planetary microbiome structure and generalist-driven gene flow across disparate habitats" analysing 85k metagenomes across the world, e.g. looking into generalism across habitats www.biorxiv.org/content/10.1... – see the thread below for more details!
#MicroSky 🖥️🧬🦠
21.07.2025 13:57 — 👍 24 🔁 12 💬 0 📌 0

Terrestrial Ecology
The department of Terrestrial Ecology studies species, communities & ecosystems in a rapidly changing world.
I am so excited to share that I will be joining the Terrestrial Ecology Department at the Netherlands Institute of Ecology (NIOO-KNAW) as a tenure track senior scientist this Fall! Impossible without the unwavering support and mentorship of Jim Bever, Peggy Schutlz, Evan Gora, and Maggie Wagner.
18.07.2025 06:07 — 👍 54 🔁 4 💬 6 📌 0


BugBuster: a novel automatic and reproducible [Nextflow] workflow for metagenomic data analysis academic.oup.com/bioinformati... 🧬🖥️🧪 github.com/gene2dis/Bug...
15.07.2025 15:30 — 👍 4 🔁 3 💬 0 📌 0
🔬 24-month postdoc position Opening: "Rice Disease Metagenomics in East Africa"
Join UMR PHIM (Montpellier, France) @phimresearch.bsky.social
🧬 Oxford Nanopore long-read metagenomics
🌍 3-month field mission in Kenya
⚙️ Recent PhDs
Apply: sebastien.cunnac@ird.fr
#PostdocJobs #Metagenomics #Rice
09.07.2025 15:35 — 👍 2 🔁 3 💬 0 📌 0
🇳🇱 Galaxy is now available in the SURF Research Cloud.
Dutch researchers can launch Galaxy workspaces with configurable resources, data federation, and support for Pulsar, all in a secure, reproducible environment.
galaxyproject.org/news/2025-07...
09.07.2025 12:24 — 👍 0 🔁 2 💬 1 📌 0
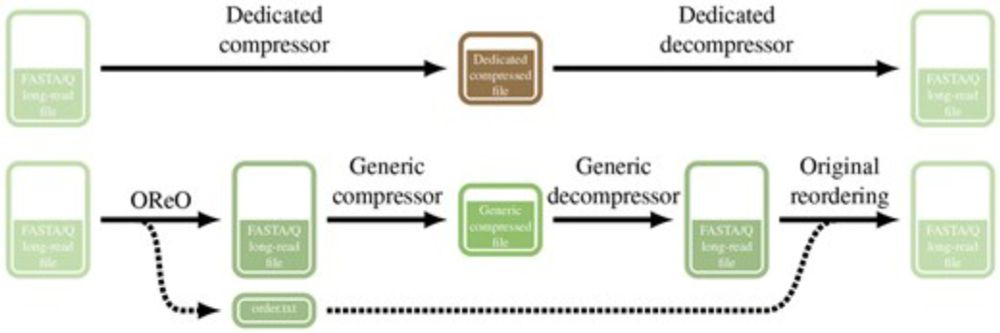
OReO: optimizing read order for practical compression
AbstractMotivation. Recent advances in high-throughput and third-generation sequencing technologies have created significant challenges in storing and mana
Paper alert!
We present Oreo a tools that reorder long reads datasets in a way to compress them efficiently with ANY universal compressor like gz, zstd, xz ...
TLDR: You can get state of the art compression WITHOUT a dedicated compressor/decompressor!
academic.oup.com/bioinformati...
A thread!
03.07.2025 10:52 — 👍 23 🔁 18 💬 1 📌 1
The team's first preprint is out!
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
27.06.2025 20:09 — 👍 46 🔁 30 💬 1 📌 3
New preprint! Deacon is a versatile tool for filtering FASTA/FASTQ files and streams at hundreds of megabases per second using minimizers, built with rapid metagenomic host depletion in mind, but equally useful for search.
github.com/bede/deacon
13.06.2025 13:24 — 👍 64 🔁 42 💬 5 📌 2
📢Our synthesis of current knowledge on the fascinating cell of #arbuscular #mycorrhizal fungi is out @currentbiology.bsky.social, part of the special #fungi issue. With @rachaelcargill.bsky.social @tobykiers.bsky.social @thomasshimizu.bsky.social
Open access link: www.cell.com/current-biol...
1/4
10.06.2025 09:20 — 👍 59 🔁 27 💬 3 📌 2

Have you ever heard of the term “dark taxa”?
Published in @currentbiology.bsky.social, a new study reveals an astonishing 83% of ectomycorrhizal #fungi are classified as dark taxa, existing only through their DNA sequences and eluding formal classification. 🧵
11.06.2025 11:56 — 👍 22 🔁 10 💬 2 📌 2
home | GlobDB
I'm happy to announce the latest release of the GlobDB, available at globdb.org.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
10.06.2025 11:20 — 👍 114 🔁 62 💬 3 📌 4
I am happy to see that this beautiful review about detecting rare organisms in metagenomics is now available: kwnsfk27.r.eu-west-1.awstrack.me/L0/https:%2F... congrats Marios Valmas and Sotirios Vasileiadis for leading and doing the heavy lifting!
08.06.2025 08:43 — 👍 3 🔁 1 💬 0 📌 0

Excited to share the #Snakemake monitoring tool I've been working on:
snkmt (Snakemate) - a CLI utility for real-time workflow monitoring. Basically solves the "is my pipeline still running?" problem and makes debugging failed jobs much easier.
#bioinformatics #workflow #opensource 1/3
03.06.2025 14:52 — 👍 13 🔁 4 💬 1 📌 0
Promising toolkit for binning and visualizing assembly graphs. The assembly graph is an underutilized source of binning information IMO
gbintk.readthedocs.io/en/latest/
30.05.2025 02:51 — 👍 52 🔁 27 💬 1 📌 0
myloasm - metagenomic assembly with (noisy) long reads
Announcing myloasm, a new long-read (ONT R10/PacBio) metagenome assembler that I've been working on during my postdoc in the Heng Li lab (@lh3lh3.bsky.social).
myloasm-docs.github.io
28.05.2025 17:53 — 👍 132 🔁 78 💬 5 📌 3
These boxes are not moving. A mind-bending optical illusion by Japanese artist Jagarikin.
07.05.2025 01:03 — 👍 9390 🔁 2370 💬 265 📌 329
What percent reads mapped to MAGs is "good" for soil or sediment samples?
is lower than water samples OK/expected due to more complex communities?
🙏
🖥️🧬 microbiome, metagenomics
06.05.2025 16:27 — 👍 13 🔁 7 💬 5 📌 0
Marie Curie Staff Exchanges Program (Nº 101130685), 2024-2028 🇪🇸 🇬🇷 🇮🇹 🇫🇷 🇳🇱 🇹🇭
Official account (for updates about PMS2025 https://6thplantmicrobiomesymposium2025.com)
PhD candidate researching Plant-microbe Interactions, Microbial genomics and Ecology @ Institute of Biology Leiden 🇳🇱 & IHSM Malaga 🇪🇸
PhD, He/Him, 🦀
- Bioinformatician, Machine Learning using Rust and R.
- I don’t use and l don’t like LLM.
- Typos in posts are meaningless auto-correct.
- Sports Model Trainer
- Read fake replies i suffered and don't support that👇 fake account and people
Welcome to the real world. Prik door de waan van de dag en zie wat er schuilgaat achter de ontwikkelingen van vandaag.
https://t.mtrbio.com/tegenlicht
An open source platform for sustainable data science. Join us at https://renkulab.io.
Built with ❤️ in 🇨🇭 at ETHZ and EPFL.
Also at https://fosstodon.org/@renku
We make free, open-source software for data scientists like the RStudio IDE.
We're formerly known as RStudio. You can always download our open-source IDE here. https://posit.co/download/rstudio-desktop/
Computational biologist and microbial ecologist at NIOO-KNAW, advancing ecosystem resilience through integrated research on plant-microbe interactions and the biodiversity of unique systems like the Galápagos. Committed to interdisciplinary collaboration.
Senior Researcher at the Netherlands Institute of Ecology (NIOO-KNAW) focused on plant-microbe interactions and drivers of microbial and plant biogeography and ecology
👨💻 @Nextflow.io and @nf-co.re Senior Developer Advocate at @Seqera.io 🇪🇸
👨🔬 Former Institut Curie & @Sorbonne-universite.fr 🇫🇷
👨🎓 Former LAIS / HUOL & BioME - UFRN 🇧🇷.
The leading provider of open source workflow orchestration software for data pipelines, cloud infrastructure & collaboration. The creators of @nextflow.io and @multiqc.info
Scientist.
Pangenomes, symbiosis, evolution, metagenomics.
MicroHealth consortium.
Postdoc University of Amsterdam; previously NIOZ 🇳🇱; EMBL🇩🇪; North Dakota State University🇺🇲, Fulbright Program; Taras Shevchenko National University of Kyiv, Ukraine🇺🇦
MultiXscale is a EuroHPC JU Centre of Excellence in multiscale modelling. It is a collaborative 4-year project between the CECAM network and EESSI.
European Environment for Scientific Software Installations (EESSI, pronounced as "easy") - https://eessi.io - HPCWire Reader’s Choice Award 2024 - Part of EuroHPC Federation Platform as Federated Software Component
Computational microbiologist. Senior scientist at @cemess.bsky.social, @univie.ac.at.
Microbial ecology, mostly of nitrogen cycle microbes, and data driven physiology.
Maintainer of the GlobDB genome database https://globdb.org
Prof. Microbiology @ Freie Universität Berlin (He/ Him)
Microbial ecology, plant leaf (phyllosphere) microbiology, fond of expressing fluorescent proteins in uncommon bacteria. Recently, we have adopted butterflies and their offspring in our lab.
Researcher at the Netherlands Institute of Ecology: plant adaptation, ecological genetics & epigenetics, molecular ecology
Located at the University of California-Berkeley at the Innovative Genomics Institute.
Purveyors of Microbial Ecology, Bioinformatics, & Nanogeoscience.
Reposts or likes≠endorsements.
https://www.banfieldlab.com/
Developing open source tools for collaborative data science at the Swiss Data Science Center.
https://renkulab.io
Computational microbiologist
I like to post about: microbial genomics, microbial ecology, evolution, micro+plant biotechnology, climate, symbiosis, virology, ag, sci publishing and policy

















