
For more information, check the preprint! (9/9)
www.biorxiv.org/content/10.1101/2024.11.14.623630v1
@elanasimon.bsky.social

For more information, check the preprint! (9/9)
www.biorxiv.org/content/10.1101/2024.11.14.623630v1

🛠️ Want to analyze your own protein models? (8/9)
- Code: github.com/ElanaPearl/interPLM
- Full framework for PLM interpretation
- Methods for training, analysis, and visualization
✨ Explore the features yourself! (7/9)
- Interactive visualization: interplm.ai
- Explore features from every layer of ESM-2-8M
- See how proteins activate different features
- Examine structural patterns

🧪 We can also steer model predictions by adjusting feature activations, demonstrating how understanding these representations could help guide protein design (6/9)
19.11.2024 19:34 — 👍 0 🔁 0 💬 1 📌 0
🎯 Beyond understanding PLMs, these features have practical applications (5/9):
Finding missing annotations in protein databases
Identifying potentially new biological motifs
Suggesting locations of binding sites and functional regions

🤖 We showed LLMs can generate meaningful descriptions of many features - and these descriptions can be validated by successfully predicted which proteins would activate each feature! (4/9)
19.11.2024 19:34 — 👍 0 🔁 0 💬 1 📌 0
📊We identified up to 2,548 interpretable features per layer that match known biological concept annotations - compared to just 46 from individual neurons.
This suggests PLMs store biological information in superposition - multiple concepts sharing the same neurons! (3/9)

🔍 Using InterPLM, we identified features in ESM-2 that detect various biological properties, from local motifs to complex structural patterns (2/9)
- Catalytic sites
- Zinc fingers
- Targeting sequences
- Post-translational modifications
- Structural elements and many more!

🧬 What are protein language models (PLMs) actually learning about biology? Our paper introduces InterPLM - a framework that reveals interpretable features in PLMs using sparse autoencoders, giving us a window into how these models represent protein structure and function.
🧵(1/8)

www.biorxiv.org/content/10.1...
InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders
Code: github.com/ElanaPearl/I...
Interactive site: interplm.ai
Nice work by Elana Simon from James Zou lab
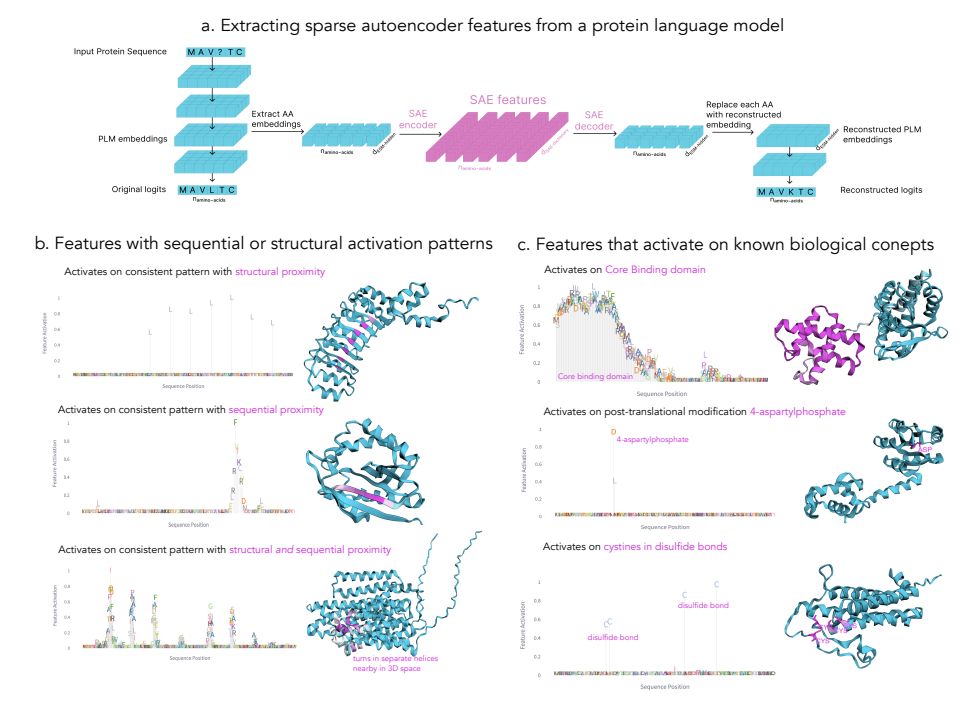
Overview of SAE methodology and representative SAE features revealed through automated activation pattern analysis
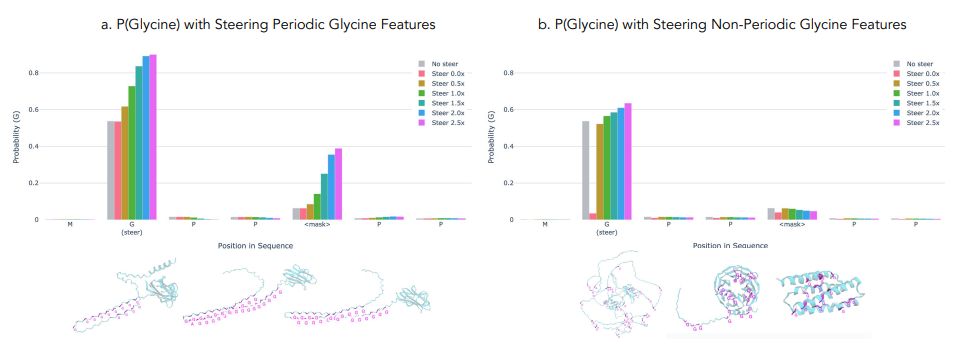
Using mechanistic interpretability to steer generations
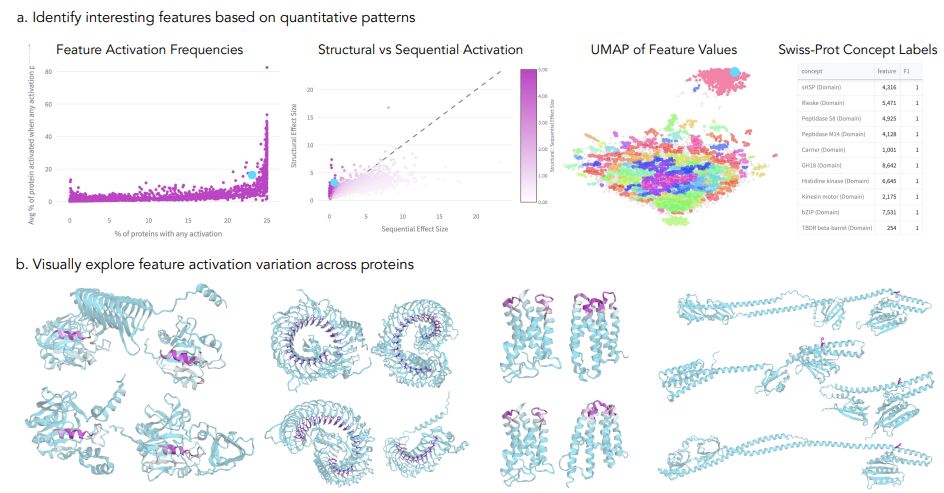
SAE feature analysis and visualizations reveal features with diverse and consistent activation patterns
Mechanistic interpretability on a protein language model
www.biorxiv.org/content/10.1...