



Second day of AITHYRA Symposium #AIforLifeScience: Cracking the Code—Genomics, Chromatin and RNA Regulation
09.09.2025 09:36 — 👍 12 🔁 2 💬 0 📌 0@lucanaef.bsky.social
Co-Founder & CTO, @vant_ai Forbes 30u30 but won't go to jail | prev. ETH Zurich Stanford McKinsey AI x Bio/Chem. vi/vim




Second day of AITHYRA Symposium #AIforLifeScience: Cracking the Code—Genomics, Chromatin and RNA Regulation
09.09.2025 09:36 — 👍 12 🔁 2 💬 0 📌 0
We have multiple senior scientist / engineer positions available across several geos in MSR AI for Science. Ping me or any of my colleagues if you have any questions!
www.microsoft.com/en-us/resear...

@aithyra.bsky.social Opening Symposium "AI for Life Science" with Nobel Laureate Frances Arnold as keynote speaker in addition to an outstanding line up of speakers on a variety of topics across biological scales and data modalities.
Registration is now open at cemm.at/aithyra-symp...
BioEmu now published in @science.org !!
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
Hello, we are AITHYRA!
I am very excited to share with you the corporate identity of AITHYRA, the Research Institute for Biomedical AI.
Check out the brand design video (sound on!) to learn more.
**REMINDER**
One week left to apply for Starting PI positions (Life Science and AI/ML). Come join us!
it's a tongue-in-cheek comment on the quote from Demis. W/ current methods, data availability is the primary determinant of performance (see plot), and unlike quote, DM hasn't found a magic "innovative algorithm" to get around it. I think both more (diverse) data + better algos will be part of soln
08.02.2025 22:44 — 👍 5 🔁 0 💬 0 📌 0lmao
08.02.2025 15:03 — 👍 3 🔁 0 💬 0 📌 0Note that I believe that still much improvement can & will be made on model side & even more so, leveraging more existing data sources. But it's very clear that only focusing on the model is a losing strategy with no moat.
08.02.2025 12:37 — 👍 6 🔁 0 💬 0 📌 0“I feel like they always talk about [...] we don’t have enough data [...] You do have enough data — if you were innovative enough on your algorithm side.”
didn't age well - turns out, having data is all that matters, and current models are largely equivalent. Fantastic work by the team!
This is the kind of benchmarking work the field needs!
24.12.2024 16:06 — 👍 14 🔁 3 💬 0 📌 0End of year notes from #PLINDER team!
It's been an incredible year for datasets and benchmarks for the #ML / #CompChem / #CompBio community.
Superbly grateful for the positive responses, constructive feedback and contributions!
It's just the start, lots more in 2️⃣0️⃣2️⃣5️⃣ 🎉
www.plinder.sh/blog/updates
Here's a year-end update from #PLINDER. It's been really great working on this project and all the other projects that it has kickstarted - the gift that keeps on giving.
Happy holidays everyone!
www.plinder.sh/blog/updates
🫡🚀 lets gooo
23.12.2024 19:13 — 👍 1 🔁 0 💬 0 📌 0Two major life updates:
- I'm moving to Senior Applied Research Scientist in Digital Biology at NVIDIA (Jan '25)
- I'm starting a new lab at Duke as Visiting Assistant Prof (early '25)
Both roles focus on tackling hard problems in biological machine learning through collaborative research.
Long 🧵
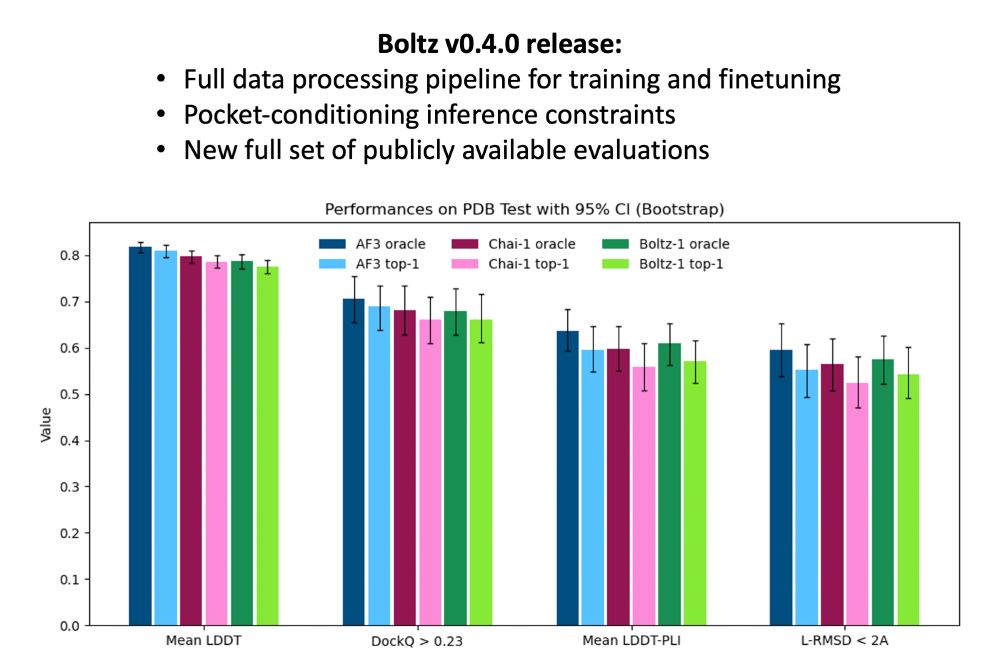
Boltz v0.4.0 is here! Today, we’re releasing our full data processing pipeline, making it easier than ever to build on top of Boltz. This release also includes our evaluation code and new results. Oh, and also pocket conditioning :)
21.12.2024 16:26 — 👍 60 🔁 15 💬 1 📌 0
Bonus: you can now watch on YouTube the recording of the seminar we gave on Boltz-1 with @mmbronstein.bsky.social and VantAI! www.youtube.com/watch?v=mN5p...
21.12.2024 17:08 — 👍 30 🔁 8 💬 0 📌 0
FutureHouse is launching an independent postdoctoral fellowship program for exceptional researchers who want to apply our automated science tools to specific problems in biology and biochemistry, in collaboration with world-leading academic labs. 1/
19.12.2024 15:29 — 👍 49 🔁 25 💬 1 📌 9actually agree here that we should provide "sensible defaults". I think we could just provide a few presets ("high quality balanced", "high-data balanced", "unfiltered-unbalanced"). On the exact specifics of those presets would be great to share notes, you have a lot of exp here obviously
16.12.2024 18:17 — 👍 3 🔁 0 💬 1 📌 0but if we can make metrics less strict that would give leeway to optimize relevance so this great. By no means current splits are the final word, it's a framework to research splits easily. 💯 agree that more than 1 split is likely end-state. Would love to work together on the next iteration! (2/2)
16.12.2024 02:03 — 👍 6 🔁 0 💬 0 📌 0This is incredibly (!) useful David, really appreciate you taking the time to dig in. Orthogonal descriptors validation is great to see. We'll check what happened with redundant ligand in tests, intention was to remove that. it's non-trivial to optimize generalizability, relevance and quality (1/2)
16.12.2024 02:03 — 👍 6 🔁 0 💬 1 📌 0Congrats!! 🥳
20.11.2024 00:39 — 👍 2 🔁 0 💬 0 📌 0