
Mirdita Lab - Laboratory for Computational Biology & Molecular Machine Learning
Mirdita Lab builds scalable bioinformatics methods.
My time in @martinsteinegger.bsky.social's group is ending, but I’m staying in Korea to build a lab at Sungkyunkwan University School of Medicine. If you or someone you know is interested in molecular machine learning and open-source bioinformatics, please reach out. I am hiring!
mirdita.org
20.01.2026 11:07 — 👍 105 🔁 54 💬 7 📌 1


Just coincidentally found GenBank Release 84.0 from 1994 in the neighboring lab. Anyone out there with an even older version?
26.01.2025 02:28 — 👍 80 🔁 13 💬 6 📌 2
In case you missed our ML for proteins seminar on CHEAP compression for protein embeddings back in October, here it is -- thanks @megthescientist.bsky.social for doing so much for the MLxProteins community 🫶
28.12.2024 03:00 — 👍 16 🔁 1 💬 2 📌 0

•introduced “zero shot prediction” as a question of guessing a bioassay’s outcome by likelihoods of pLMs
•commented on biases in evolutionary signals from Tree of life used to train pLMs (a favorite paper I read in 2024: shorturl.at/fbC7g)
16.12.2024 06:29 — 👍 8 🔁 1 💬 1 📌 0
Thanks @workshopmlsb.bsky.social for letting us share our work!
🔗📄 bit.ly/plaid-proteins
15.12.2024 22:27 — 👍 22 🔁 2 💬 1 📌 0
Another straightforward application is generation, either by next-token sampling or MaskGIT style denoising. We made the tokenized version of CHEAP to do generation, and decided to go with diffusion on continuous embeddings instead — but I think either would’ve worked
10.12.2024 01:04 — 👍 6 🔁 0 💬 0 📌 0
We trained a model to co-generate protein sequence and structure by working in the ESMFold latent space, which encodes both. PLAID only requires sequences for training but generates all-atom structures!
Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!
09.12.2024 14:58 — 👍 57 🔁 11 💬 2 📌 0
immensely grateful for awesome collaborators on this work: Wilson Yan, Sarah Robinson, @kevinkaichuang.bsky.social, Vladimir Gligorijevic, @kyunghyuncho.bsky.social, Rich Bonneau, Pieter Abbeel, @ncfrey.bsky.social 🫶
06.12.2024 17:44 — 👍 3 🔁 0 💬 0 📌 0
6/ We'll get to share PLAID as an oral presentation at MLSB next week 🥳 In the meantime, checkout:
📄Preprint: biorxiv.org/content/10.1...
👩💻Code: github.com/amyxlu/plaid
🏋️Weights: huggingface.co/amyxlu/plaid...
🌐Website: amyxlu.github.io/plaid/
🍦Server: coming soon!
06.12.2024 17:44 — 👍 6 🔁 1 💬 1 📌 0

conditioning on organism and function shows that PLAID has learned active site residues and sidechain positions!
5/🚀 ...and when prompted by function, PLAID learns sequence motifs at active sites & directly outputs sidechain positions, which backbone-only methods such as RFDiffusion can't do out-of-the-box.
The residues aren't directly adjacent, suggesting that the model isn't simply memorizing training data:
06.12.2024 17:44 — 👍 5 🔁 2 💬 1 📌 0

unconditional generations from PLAID
4/ On unconditional generation, PLAID generates high quality and diverse structures, especially at longer sequence lengths where previous methods underperform...
06.12.2024 17:44 — 👍 5 🔁 0 💬 1 📌 0

noising by a diffusion schedule in the latent space doesn't always correspond to the same corruption in the sequence and structure space...
3/ I was pretty stuck until building out the CHEAP (bit.ly/cheap-proteins) autoencoders that compressed & smoothed out the latent space: interestingly, gradual noise added to the ESMFold latent space doesn't actually corrupt the sequence and structure until the final forward diffusion timesteps 🤔
06.12.2024 17:44 — 👍 4 🔁 0 💬 1 📌 0

how does the PLAID approach work?
2/💡Co-generating sequence and structure is hard. A key insight is that to get embeddings of the ESMFold latent space during training, we only need sequence inputs.
For inference, we can sample latent embeddings & use frozen sequence/structure decoders to get all-atom structure:
06.12.2024 17:44 — 👍 4 🔁 0 💬 1 📌 0
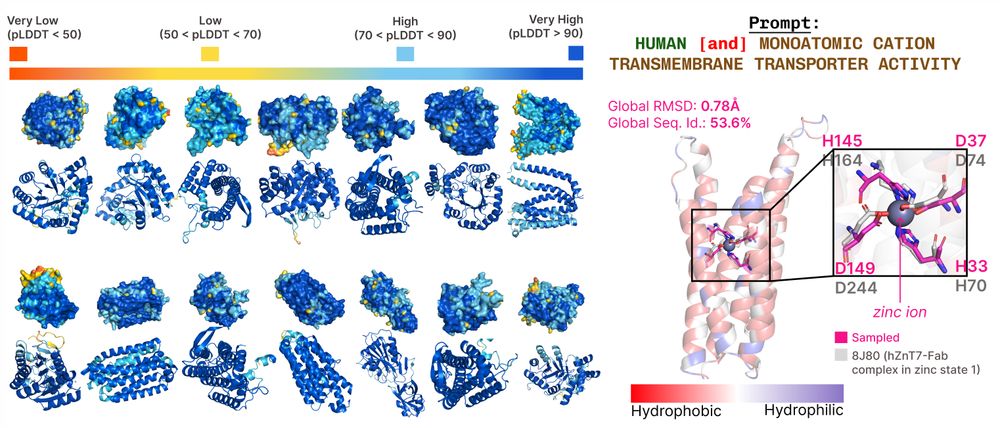
overview of results for PLAID!
1/🧬 Excited to share PLAID, our new approach for co-generating sequence and all-atom protein structures by sampling from the latent space of ESMFold. This requires only sequences during training, which unlocks more data and annotations:
bit.ly/plaid-proteins
🧵
06.12.2024 17:44 — 👍 121 🔁 37 💬 1 📌 3
biology 🧬, microscopes 🔬, and computers 🖥️. Preferably all at once.
this bio is left as an exercise to the reader
🌱 emilyliu.me
Stanford Bioengineering PhD candidate / Biological AI in Brian Hie’s lab at Arc Institute
https://samuelking.cargo.site
enjoying and bemoaning biology. phd student
@columbia prev. @harvardmed @ginkgo @yale
Research Engineer at Google DeepMind. AlphaFold, LLMs, Physics and Civic Tech. tfgg.me
Learning from algorithms that learn from data about gene regulation.
Founder/CEO @ AINovo Biotech. Prev: AI + Genomics @ Stanford Computer Science, computational drug discovery @ ETH, Harvard. AI Optimist for Human Health.
phd student @ berkeley ai, vision + language
🔗 https://people.eecs.berkeley.edu/~graceluo
🐦 https://x.com/graceluo_
Scientist, #MachineLearning and #AI for Moleculear Sciences. Scuba Diver. Loves @cecclementi.bsky.social
Research Scientist @prescientdesign @Genentech Former PhD @umdcs
ayaismail.com
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan
Previously IMP Vienna, Stanford Genetics, UW CSE.
Doudna Lab at UC Berkeley, Innovative Genomics Institute founder, CRISPR co-inventor and Nobel laureate innovativegenomics.org/
@_angie_chen at the other place
PhD student @NYU, formerly at
@Princeton 🐅
Interested in LLMs/NLP, pastries, and running. She/her.
AI researcher in music, audio, LLMs.
Bioinformatics PhD Student in the Ren lab at UCSD. Modeling gene regulation across species.








