
🧪Scientists from our Haselbach lab captured how proteins begin to fold as they’re being made.
Using cryo-EM, they visualised chaperones guiding nascent proteins on the ribosome: https://www.nature.com/articles/s41467-025-67685-6
@paloha.bsky.social
Computational Scientist for BioAI & Cryo-Electron Tomography @ ISTA | Co-Founder of https://ACAI.AI.

🧪Scientists from our Haselbach lab captured how proteins begin to fold as they’re being made.
Using cryo-EM, they visualised chaperones guiding nascent proteins on the ribosome: https://www.nature.com/articles/s41467-025-67685-6
New publication by ISTA scientists @aliciakmichael.bsky.social, @paloha.bsky.social, and collaborators made it to @cp-molcell.bsky.social's cover. The study demonstrates the potential of modern cryo-ET techniques and open data sharing to empower visual proteomics. More: bit.ly/49wzHdr
14.01.2026 10:27 — 👍 5 🔁 2 💬 1 📌 0This one was quite the journey! The paper describing the #ChlamyDataset is finally out and on the cover of Mol Cell!
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]

Check also the table and star files in github.com/Chromatin-St...
28.12.2025 09:49 — 👍 2 🔁 0 💬 0 📌 0
Online Now: Toward community-driven visual proteomics with large-scale cryo-electron tomography of Chlamydomonas reinhardtii Online now:
24.12.2025 20:14 — 👍 27 🔁 11 💬 0 📌 0To be fair, beautiful images from the whole team made it easier to impress, and luckily ISTA IT had no trouble with my odd printing request 🦾
Team: @janstevens.bsky.social, Volodymyr Masalitin, @ma3ke.bsky.social, @alex-bronstein.bsky.social, @cg-martini.bsky.social, @aliciakmichael.bsky.social

Photo credit: ISTA Comms & Event team

Photo credit: ISTA Comms & Event team


Our poster was awarded “Best Poster” at the Institute of Science and Technology Austria PhD Welcome Poster Session 🎉 Huge thanks to everyone who stopped by to look at our 2.5-dimensional dive into AI-Ready Cryo-Electron Tomography Simulations of the Whole Cell.
27.11.2025 12:26 — 👍 6 🔁 0 💬 1 📌 1I think your best bet with sharing hi-res videos is to just share a link to YouTube, Vimeo, or even your own website where people can open it directly.
21.11.2025 08:28 — 👍 6 🔁 0 💬 0 📌 0
70th Vienna Deep Learning Meetup
Mark your calendar for our 70th Vienna Deep Learning Meetup at Magenta Telekom. We’re bringing a lineup of great talks from Muhamed Loshi (RBI) and my colleagues Jonathan Scott & Kristina Kapanova (ISTA). Join us for federated learning, agentic AI security & scientific HPC! 🚀
RSVP: lnkd.in/dV8xVSQM
Thanks to all who helped making this possible, namely @janstevens.bsky.social, Volodymyr Masalitin, @ma3ke.bsky.social , @alex-bronstein.bsky.social , @cg-martini.bsky.social , and @aliciakmichael.bsky.social.
17.11.2025 13:59 — 👍 2 🔁 0 💬 0 📌 0
AI-Ready Cryo-Electron Tomography Simulations of the Whole Cell @ Young Scientists Symposium ISTA, in Klosterneuburg on 18 November 2025
Tomorrow, on 18 Nov, I'll again give a talk at YSS'25. This time, I will present our work on AI-Ready Cryo-Electron Tomography Simulations of the Whole Cell. This is a project I’m especially fond of. I had the idea years ago, and it’s finally taking shape at @istaresearch.bsky.social & @rug.nl
17.11.2025 13:59 — 👍 7 🔁 2 💬 1 📌 0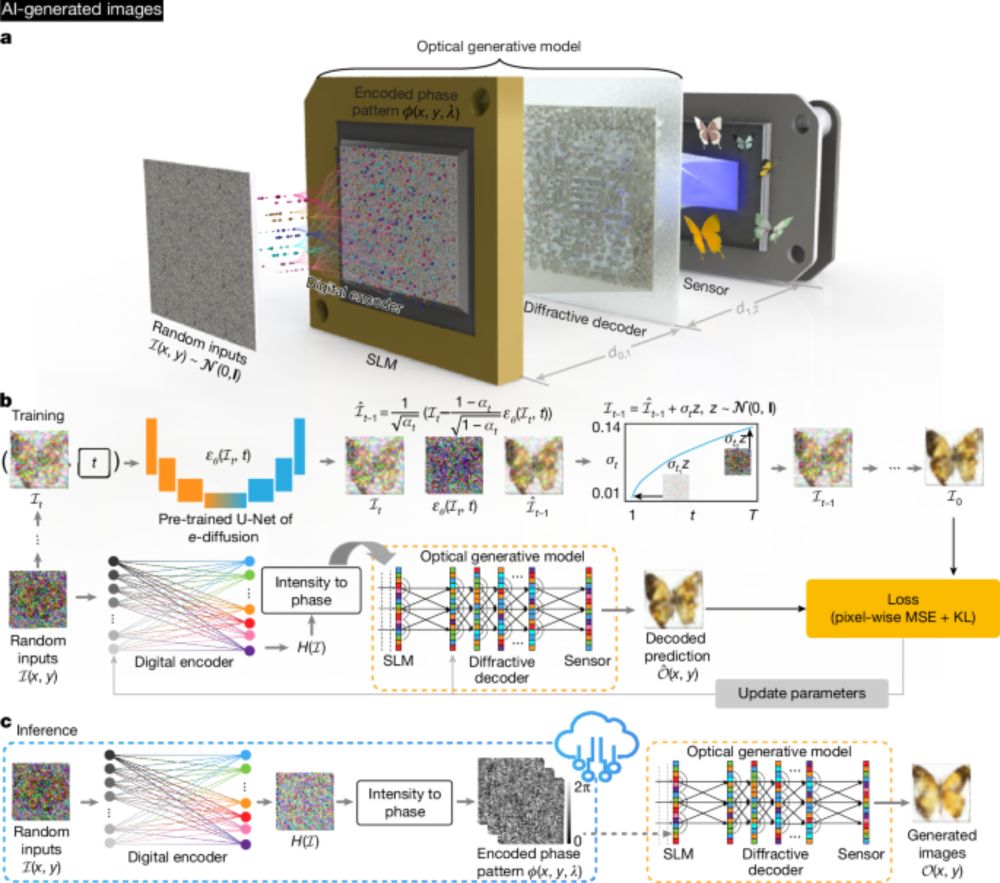
Look at this beauty. Light-based hardware for running neural networks efficiently. My totally out-of-expertise idea from years back and scientists at Bioengineering Department, UCLA are making this a reality! www.nature.com/articles/s41...
04.09.2025 12:50 — 👍 2 🔁 0 💬 0 📌 0This looks like a cowference.
15.07.2025 06:55 — 👍 2 🔁 0 💬 0 📌 0You can export One Tab links to txt.
27.05.2025 22:22 — 👍 0 🔁 0 💬 0 📌 0You might want to know about www.one-tab.com.
23.05.2025 06:48 — 👍 0 🔁 0 💬 1 📌 0Possible, let's get in touch @jni.codes. @alisterburt.bsky.social thx for the connect.
18.05.2025 19:23 — 👍 1 🔁 0 💬 0 📌 0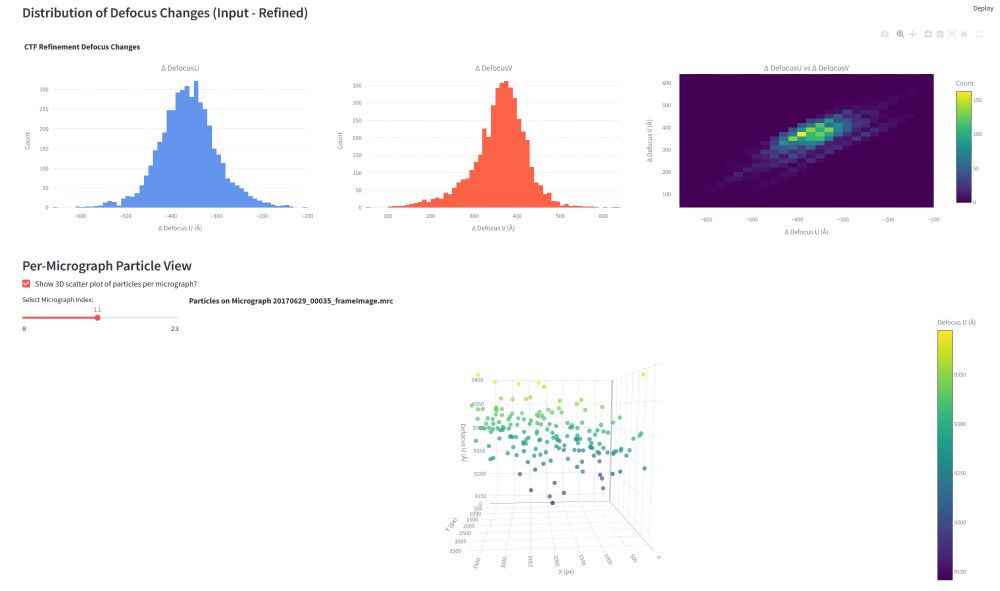
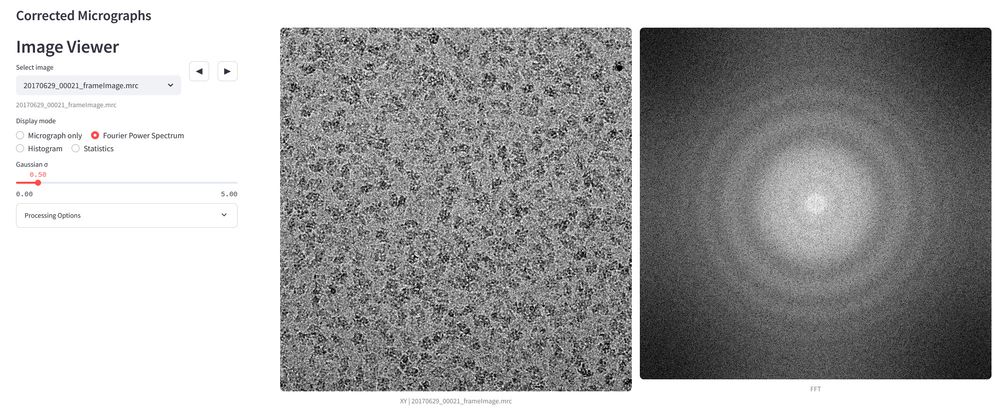
1/5: Over the last few months, I have been working on an update for the #FollowRelionGracefully dashboard, which offers improved real-time job previews for #cryoEM #Relion jobs.
27.04.2025 20:49 — 👍 50 🔁 20 💬 2 📌 2Hello, is there still space for @aliciakmichael.bsky.social & @paloha.bsky.social ? Thanks
03.04.2025 15:54 — 👍 1 🔁 0 💬 1 📌 0IMO the best invention is two-stage proposals: first short, then long, after it is selected as a good candidate. Ideally the reviewers should have an opportunity to specify what they would like to see in the longer version. So the writers can focus on answering what matters to the reviewers.
12.03.2025 11:45 — 👍 3 🔁 0 💬 0 📌 0Most often the grant agency is forcing the structure onto the writers (I believe with good intentions) rather then giving them the freedom. So as a reviewer, I'd just try to focus on fishing out the important from the suboptimal form.
12.03.2025 11:43 — 👍 3 🔁 0 💬 1 📌 0In the past years I have read many posts like this -earlier on twitter, now here, where reviewers of grants write "how they would like it". The ideas differ so much that it is impossible for anybody writing the grant to anticipate what is the best "scaffold" to follow so the reviewers are happy.
12.03.2025 08:03 — 👍 2 🔁 0 💬 1 📌 0
Let's talk about this Nature piece in more detail.
I've rarely read something so anti-scientific anywhere short of the National Review.
www.nature.com/articles/d41...
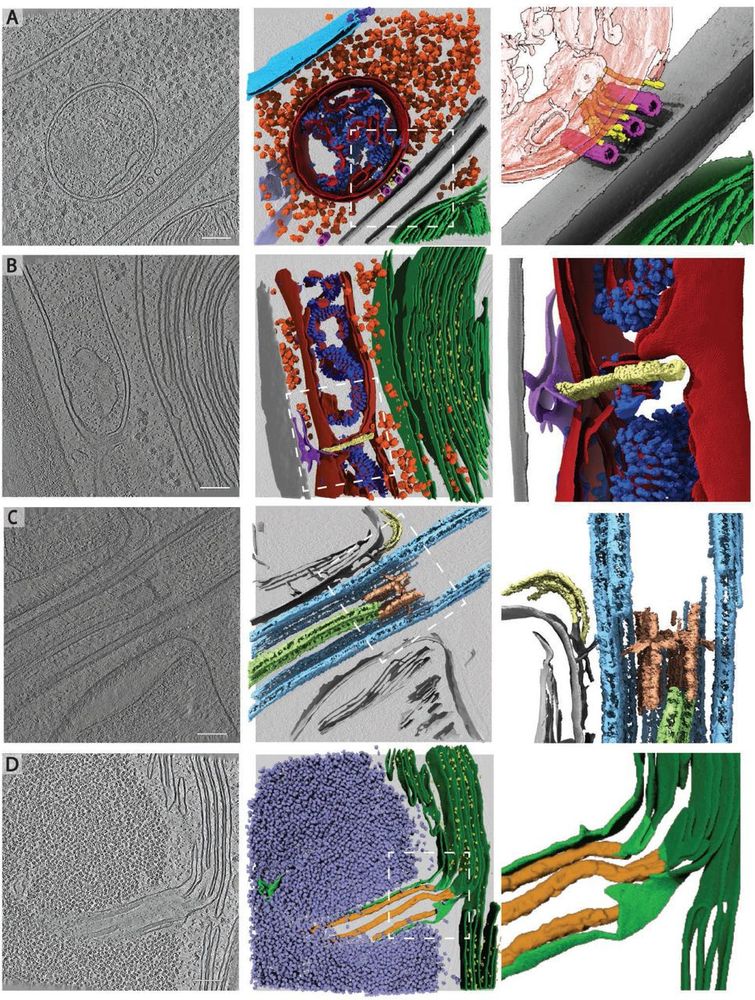
Rare cellular events observed in the cryo-ET dataset. Slices through tomograms (left) with corresponding 3D segmentation (middle and right). Right panels show enlarged views of the boxed regions in the middle panels. A) Mitochondrion at the plasma membrane, tethered to microtubules by thin filaments of unknown identity. Segmented classes: nuclear envelope (light blue), nuclear pore complex (dark blue), 80S ribosomes (orange), mitochondrial membranes (dark red), ATP synthase (royal blue), plasma membranes (grey), microtubules (pink), filaments (yellow), endoplasmic reticulum (purple). Zoomed-in panel shows thin filaments running between microtubules and the mitochondrion at the plasma membrane. Note that this tomogram contains two closely appressed cells, and thus, two plasma membranes with cell walls between. B) Mitochondrial fission event with ER membrane interactions. Segmented classes: mitochondrial membranes (dark red), ATP synthase (royal blue), 80S ribosomes (orange), thylakoid membranes (green), PSII (bright yellow), cell wall (grey), ER (purple), fission site containing filamentous structures perpendicular to the mitochondrial long axis (pale yellow). C) Ciliary transition zone between basal body and axoneme, including assembling IFT train and stellate structure77. Segmented classes: microtubule doublets (light blue), central microtubule pair (light green), stellate structure (pale red), IFT train (yellow). D) Pyrenoid tubule extending from the thylakoids into the phase-separated Rubisco matrix of the pyrenoid. Minitubules originate from thylakoid membranes82. Segmented classes: thylakoid membranes (dark green), PSII (yellow), pyrenoid tubule (lime green), Rubisco (lavender blue), minitubules (orange). Scale bars in A-D: 100 nm. Related to Fig. 2C-H.
To all #TeamTomo #CryoET and #Chlamydomonas aficionados: we have updated EMPIAR-11830 with a bug fix concerning some cryo-CARE denoised tomos as well as additional files and metadata! 🎉👩🏽💻
www.ebi.ac.uk/empiar/EMPIA...
A little thread about what's new... 🧵 1/n
Finally, my postdoc work is published in Cell Structure! 🎉 Grateful for the chance to apply deep learning to cryo-ET and learn from @haselbachlab.bsky.social about structural biology🦠. Huge thanks to Lukas Herrmann for coding help and Philipp Grohs for my position and all the GPUs.
15.02.2025 10:08 — 👍 17 🔁 3 💬 0 📌 0WOW!😍 This is exactly the type of development we hoped the Chlamy dataset would help empower! Lots of organelles and cellular features labeled automatically, context-aware particle picking, & area-selective template matching #TeamTomo 🔬
Big props to @mgflast.bsky.social @thomsharp.bsky.social 🧪🧶🧬
Thanks Alicia! I am also happy to have helped finalize this work. I hope computational biologists will find the repo useful. We need to share more annotations in accessible format, the algorithms are hungry 😄 #teamtomo, consider sharing your data via pull requests—let’s grow this together! 🤝
07.01.2025 13:18 — 👍 5 🔁 2 💬 0 📌 0This must have been a tomogram. Pre and post missing wedge correction. The elongation is uncanny.
22.11.2024 22:14 — 👍 1 🔁 0 💬 0 📌 0