
NuPhy also has some more tame keyboards, which are generally well received too. nuphy.com
26.01.2026 18:53 — 👍 1 🔁 0 💬 1 📌 0@mattbashton.bsky.social
Associate Prof in Computational Biology at Northumbria University, Newcastle, UK. Structural and High Throughout Sequencing Bioinformatics, Protein Domains, Pandemic Preparedness.

NuPhy also has some more tame keyboards, which are generally well received too. nuphy.com
26.01.2026 18:53 — 👍 1 🔁 0 💬 1 📌 0Lol just say throcky gasket in the M and S food advert voice 🤣, seriously the world of keyboards has gone crazy with levels of refinement and customisation..
26.01.2026 18:48 — 👍 1 🔁 0 💬 0 📌 0OMFG! THIS, this stupid pop-up is the bane of my existence, every bloody time I log-in, it never goes away, and the countless time's it's asked me negates any saving, I'd really like to know who is responsible for this dialogue and where I file a ticket to remove it from the universe!
26.01.2026 18:41 — 👍 1 🔁 0 💬 0 📌 0
Epomaker also has some fancy stuff epomaker.com
26.01.2026 18:36 — 👍 0 🔁 0 💬 2 📌 0
I'd have a look at both Akko en.akkogear.com and Varmilo varmilo.com things tend to be released in limited runs, with a choice of switch.
26.01.2026 18:31 — 👍 1 🔁 0 💬 2 📌 0Depends on what you want, I love my mechanical keyboards, but not everyone wants to go down that route, however the world of mechanical keyboards has vastly expanded beyond what type of cherry switch do you want.. There is now a proliferation of switches and fancy keycaps too
26.01.2026 18:31 — 👍 1 🔁 0 💬 1 📌 0This project is joint with fab computational biologist @mattbashton.bsky.social - great opportunity to learn computational and molecular biology skills
24.01.2026 15:09 — 👍 4 🔁 6 💬 0 📌 0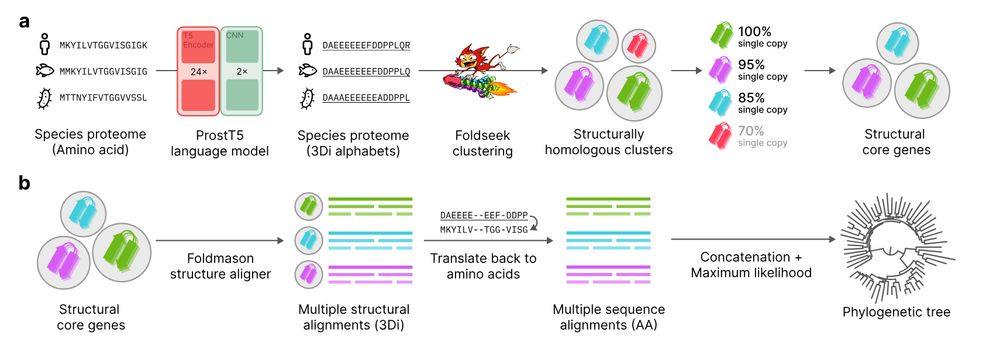
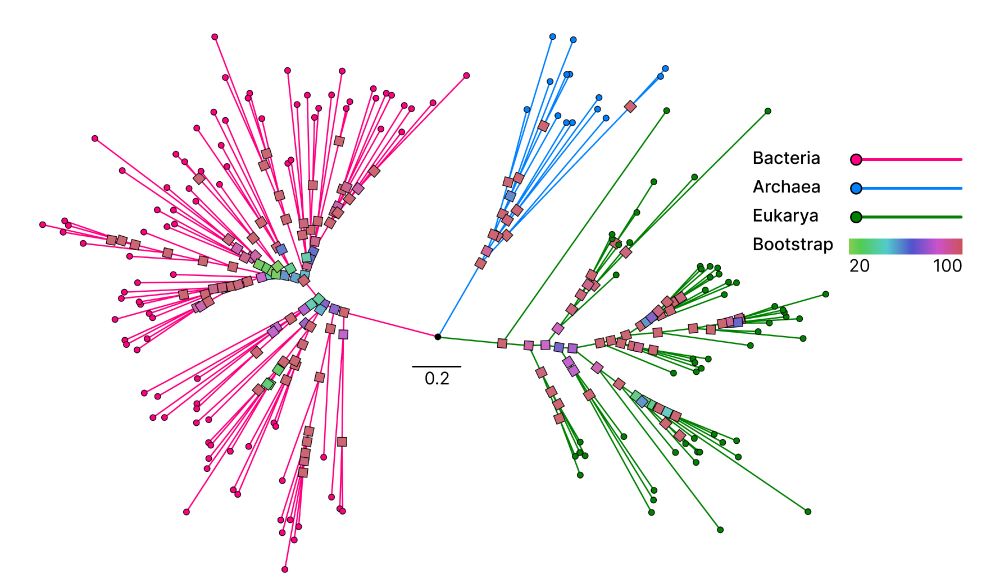
Unicore identifies single-copy protein structures across genomes using Foldseek, bypassing slow structure predictions by utilizing 3Di predictions from ProstT5, enabling rapid phylogenetic inference at the tree-of-life scale. 1/n
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
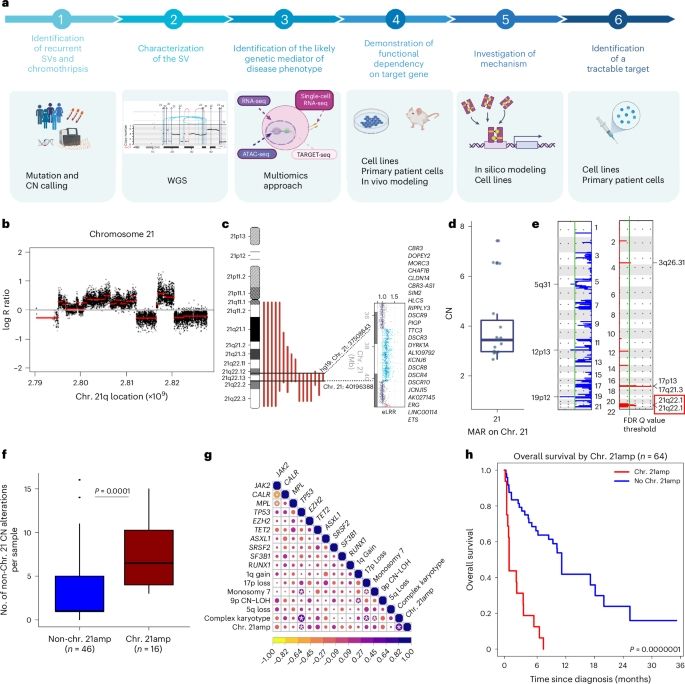
This research with Oxford started during the pandemic when I spotted a familiar iAMP21 like copy number profile on chromosome 21 here termed chr. 21amp. It's lovely to see all the different omics techniques come together from all the collaborators! www.nature.com/articles/s41...
09.06.2025 18:54 — 👍 0 🔁 0 💬 0 📌 0The age old "similar to putative function" annotation disease - only with AI, reinforcing suspect homology based matches in training data.
31.03.2025 12:05 — 👍 0 🔁 0 💬 0 📌 0Congratulations, I guess you will be spending more time in Heidelberg!
13.03.2025 19:57 — 👍 1 🔁 0 💬 0 📌 0



This week at @northumbriauni.bsky.social we launched our new #HPC cluster Higgs, having been involved in shaping this project from the outset it's nice to see it finally come to fruition with our partners #Logicalis and #Lenovo. l'm looking forward to all the research our new cluster will enable.
06.03.2025 20:43 — 👍 1 🔁 0 💬 0 📌 0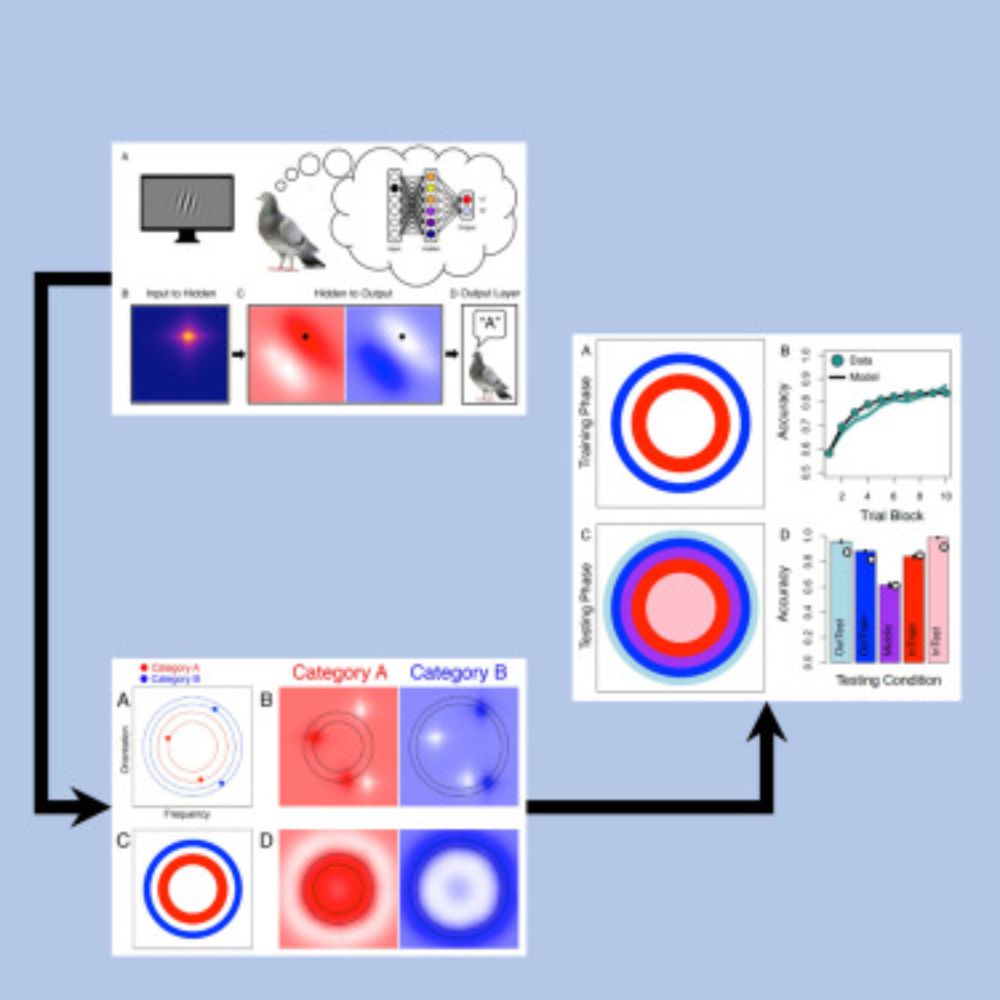
Can't afford an NVIDIA H100 just get a pigeon, readily available in all Northern towns too. cell.com/iscience/ful... #AI #Pigeon
05.03.2025 20:49 — 👍 1 🔁 0 💬 0 📌 0I'm not sure why everyone is surprised by DeepSeek if you deny people access to the latest hardware then they will naturally develop more efficient algorithms. In this sense US "export" policy on AI hardware has been a massive own goal.
29.01.2025 08:54 — 👍 6 🔁 1 💬 1 📌 0
Designers and environmental scientists from Northumbria have teamed up with The Microfibre Consortium to establish the Fibre-fragmentation and Environment Research Hub (FibER Hub) to explore the extent and environmental impact of microfibre loss from textiles
www.northumbria.ac.uk/about-us/new...

Star Trek The Next Generation scene. Early series Picard is shown up close with a determined look of contentment. He's locked into eye contact with someone just off screen, and smouldering. Closed caption reads: (champagne fizzing)
01.01.2025 04:59 — 👍 1295 🔁 270 💬 8 📌 7



Northern lights in Northumberland- beautiful and so counter intuitive that the camera is a far more faithful representation of the colours than your eyes (it’s a grey pink sky to the naked eye!)
01.01.2025 18:24 — 👍 55 🔁 7 💬 2 📌 0
and also our latest work, Natálias latest preprint, a collaboration with the groups of
@mattbashton.bsky.social and Ricardo Rajsbaum is out in BioRxiv. www.biorxiv.org/content/10.1...
I didn't have time to stop to play it, but it looks authentic, around the time it was current there were a lot of clone JAMA boards from Taiwan which had all sorts of unique enhancements like changing fighter mid match! Only they had 3x the PCB board count as reverse engineered all custom chips.
21.11.2024 15:32 — 👍 1 🔁 0 💬 1 📌 0


 20.11.2024 22:09 — 👍 1 🔁 0 💬 0 📌 0
20.11.2024 22:09 — 👍 1 🔁 0 💬 0 📌 0




Been exploring some of the breathtaking John Portman architecture in Atlanta while at #SC24
20.11.2024 21:38 — 👍 5 🔁 0 💬 1 📌 0
Nice to see this CPS-1, duel CPU 68000/Z80 set-up at #SC24, it's all about those custom chips too! #retrogaming
20.11.2024 21:32 — 👍 29 🔁 2 💬 1 📌 0
So #SC24 has been a lot of fun! We are also entering a new phase of HPC at @northumbriauni.bsky.social as we expand facilities for our researchers www.northumbria.ac.uk/about-us/new...
20.11.2024 20:38 — 👍 3 🔁 0 💬 0 📌 0who knew it would be easier to get everyone to switch social media platforms than getting them to use hg38
14.11.2024 18:45 — 👍 254 🔁 62 💬 15 📌 13
If you find this work interesting I'm currently looking for a new PhD student, see the Add here www.findaphd.com/phds/project...
14.11.2024 19:56 — 👍 1 🔁 2 💬 0 📌 0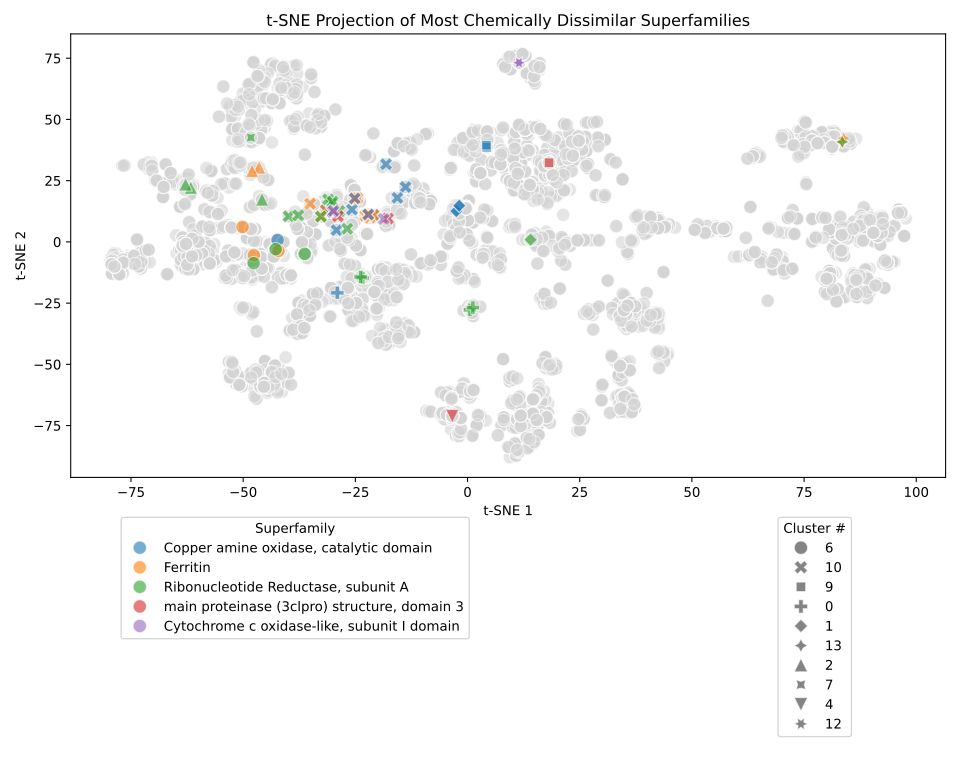
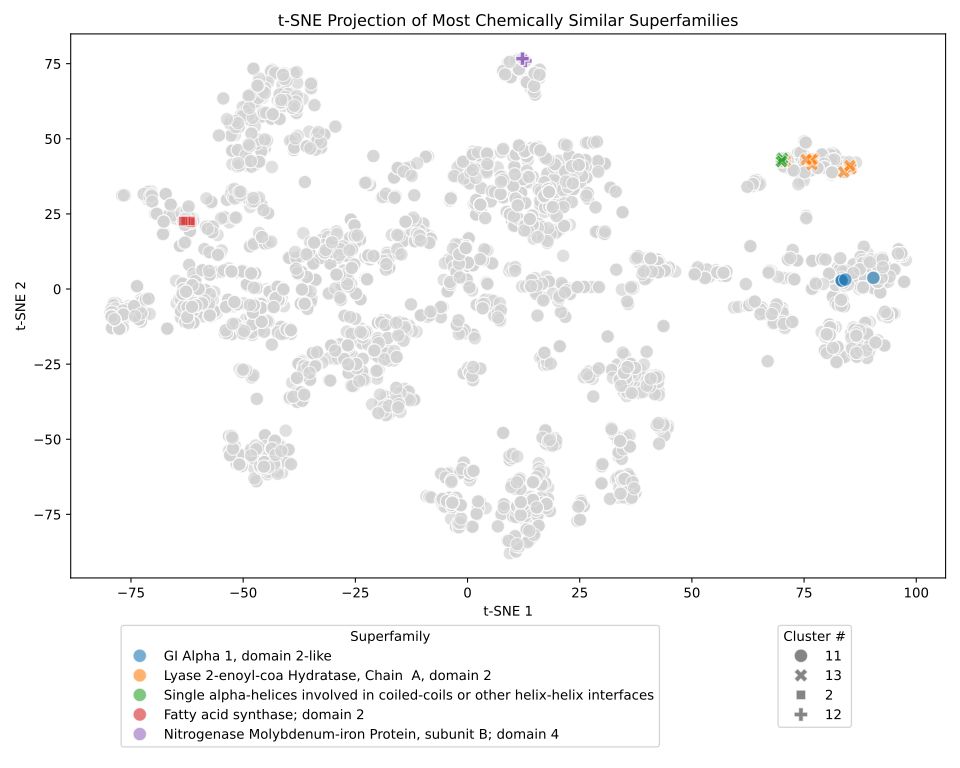
Interestingly if we look at promiscuous superfamilies' (those binding multiple ligands) we find they can fall into "generalised" - binding a wide range of chemically diverse ligands or "specialised" - binding some very chemically similar ligands, categories.
14.11.2024 19:56 — 👍 2 🔁 1 💬 1 📌 0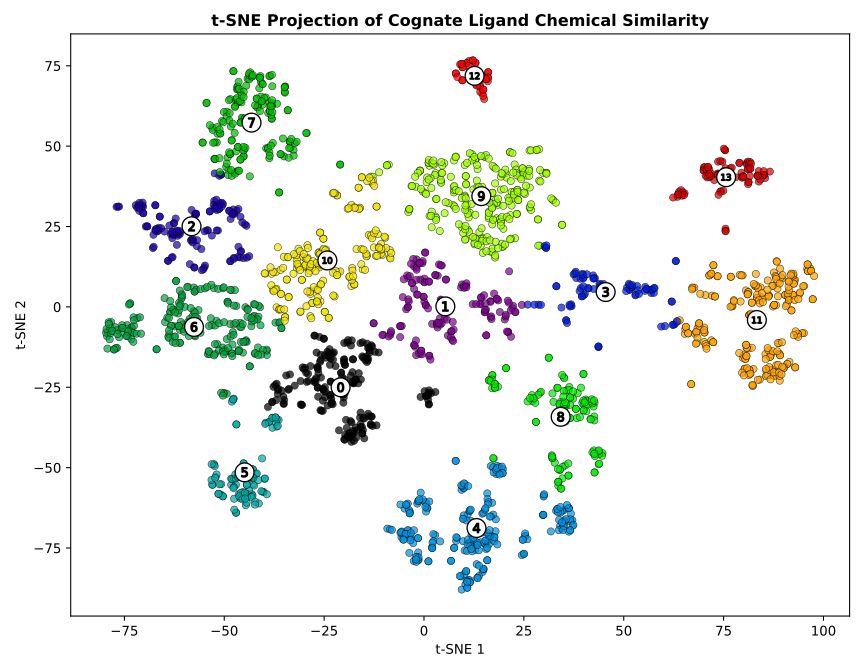
This the work of my PhD student Matt Crown, one of the nice things he did was this t-SNE plot of cognate ligand space within the PDB, in which we see some nice clusters of related compounds.
14.11.2024 19:56 — 👍 0 🔁 0 💬 1 📌 0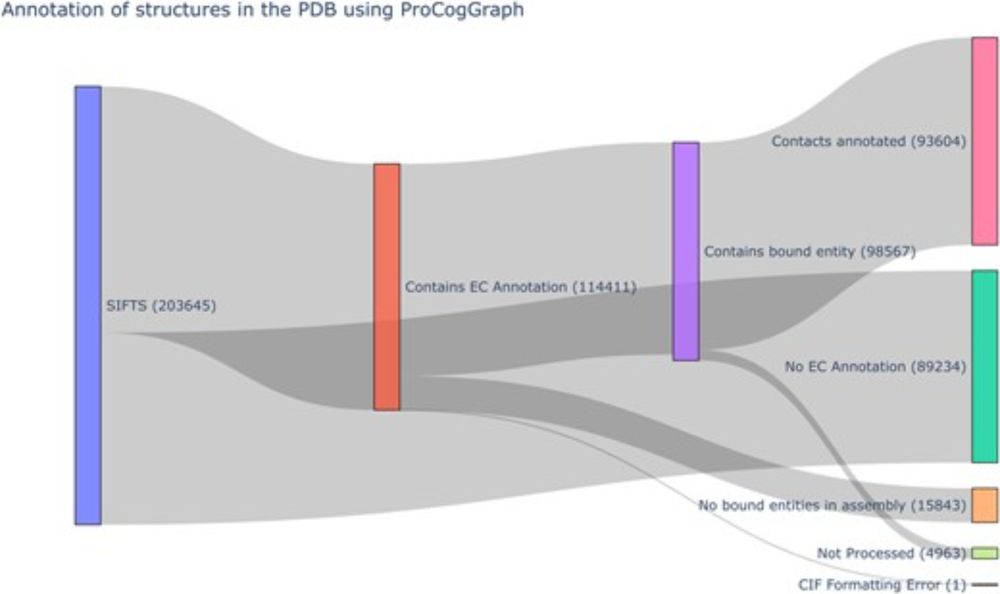
Our latest work has been published in #Bioinformatics Advances ProCogGraph, the spiritual successor of PROCOGNATE. This graph database provides a linkage between proteins domains (SCOP/CATH/Pfam) and cognate ligands for #enzymes in the #PDB. academic.oup.com/bioinformati...
14.11.2024 19:56 — 👍 4 🔁 2 💬 1 📌 0
I'm advertising a PhD studentship (open to international students) working on long read pangenome / assembly algorithms in bacteria. Would suit someone with maths/compsci/coding background. Aptitude + interest more important than experience.
Details here:
www.findaphd.com/phds/project...

After editing, this very short letter to New Scientist is even shorter now. But I think it gets the point across! @faecalmatters.bsky.social
11.11.2024 22:23 — 👍 120 🔁 34 💬 10 📌 4