why would I use snakemake when I can simply write a python script to write 51 job scripts and then write a for loop in bash to submit them?
11.11.2025 21:07 — 👍 1 🔁 0 💬 0 📌 0


🧬 🦠 🏙️
Urban vs rural lifestyles create dramatically different gut microbiomes. But how do these different gut microbiomes affect the host?
Excited to share our new paper: www.biorxiv.org/content/10.1...
05.11.2025 02:04 — 👍 35 🔁 12 💬 2 📌 0
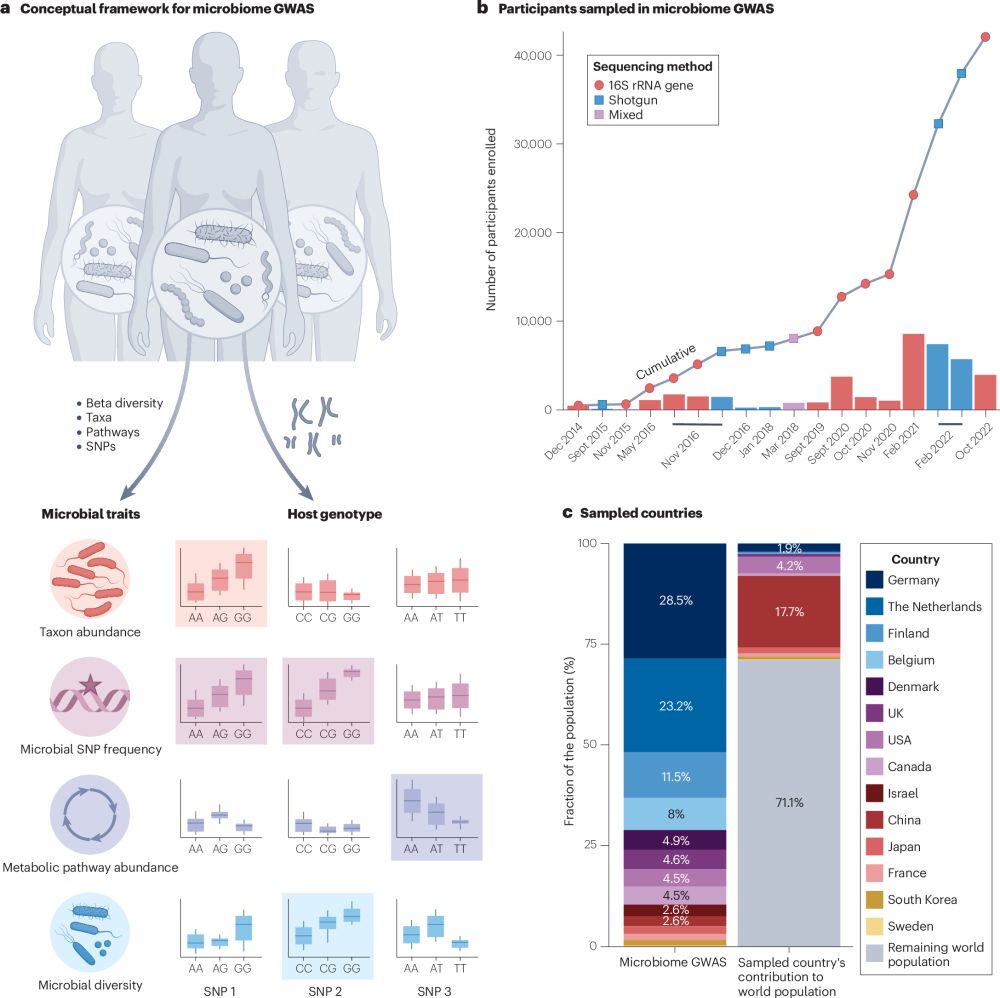
Genomics of host–microbiome interactions in humans
Nature Reviews Genetics - In this Review, Ferretti et al. discuss advances in our understanding of interactions between the human genome and the microbiome, including the effects of the microbiome...
Interested in microbiome GWAS and heritability studies? Check out our new Review in Nature Reviews Genetics! We explore key findings, challenges, and future directions of the field.
rdcu.be/epoRR
@blekhman.bsky.social @sambhawa.bsky.social and Dr. Kelsey Johnson.
04.06.2025 16:52 — 👍 48 🔁 27 💬 3 📌 3

Compendium Manager: a tool for coordination of workflow management instances for bulk data processing in Python
Compendium Manager is a command-line tool written in Python to automate the provisioning, launch, and evaluation of bioinformatics pipelines. Although workflow management tools such as Snakemake and N...
We wrote up the process we've developed for processing microbiome data in bulk! Workflow management tools are miraculous for processing a project with lots of samples, but when you have lots of *projects* too, as we do when pulling data from NCBI databases, it gets hard to juggle. #microbiomesky
19.05.2025 19:40 — 👍 7 🔁 5 💬 1 📌 1

Last week @richabdill.com and @samanthagraham.bsky.social's paper was featured on the cover of Cell!
The cover, portraying microbes on shelves shaped like a world map, was possible thanks to the amazing team of illustrators at SciStories ✏️🦠
26.02.2025 03:37 — 👍 13 🔁 6 💬 1 📌 0

These are challenging times for science, but I'm happy to share a bright moment: our work is on the cover of Cell today.
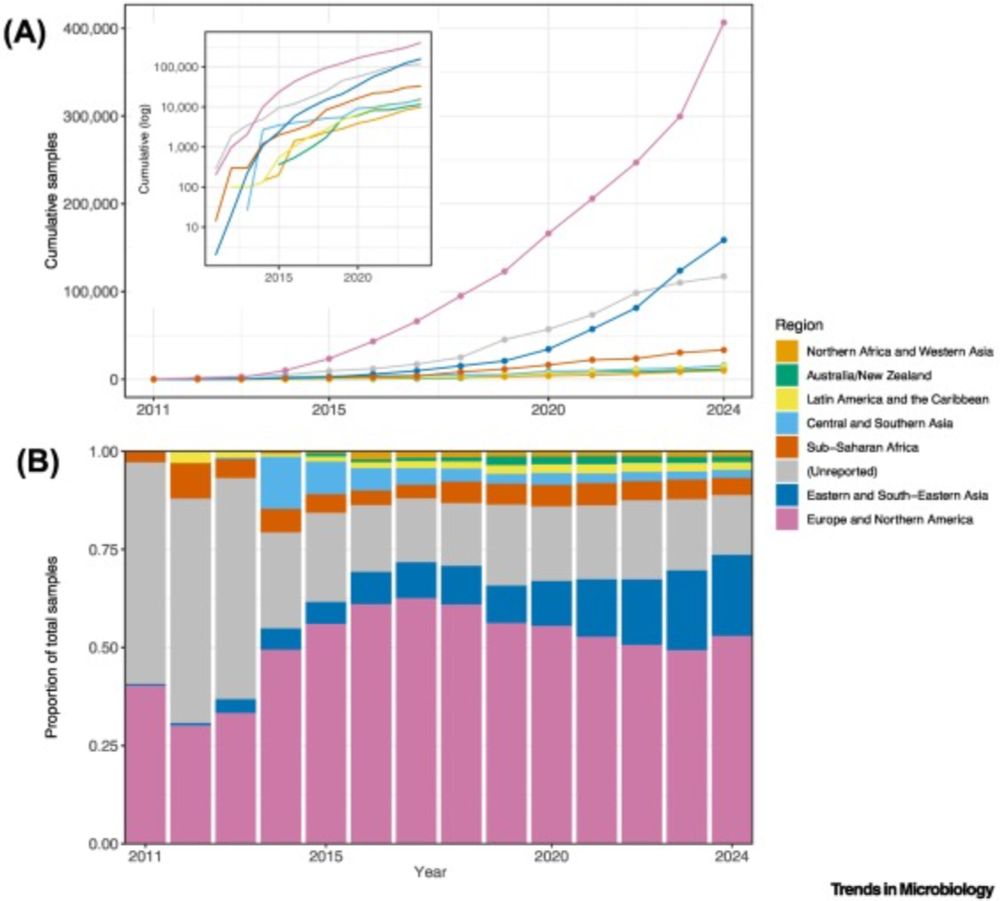
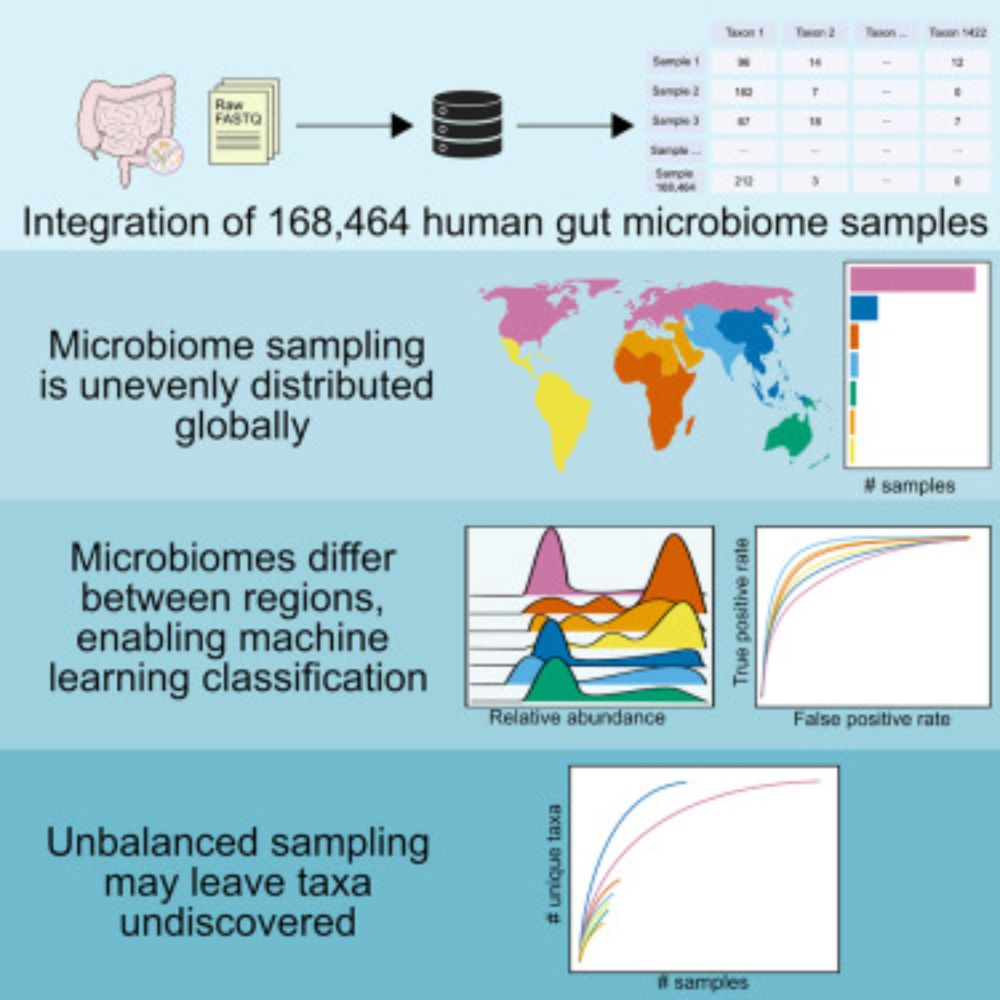
We built a compendium of human gut microbiomes integrating 168K worldwide samples, revealing patterns of microbiome variation across the globe
www.cell.com/cell/current...
20.02.2025 16:06 — 👍 143 🔁 26 💬 4 📌 0
Use the Human Microbiome Compendium data!
We made the data and code available, & created a website and R package to navigate the compendium. Links:
- Website: microbiomap.org
- Data: zenodo.org/records/1373...
- R package: blekhmanlab.github.io/MicroBioMap/
- Code: github.com/blekhmanlab/...
24.01.2025 02:50 — 👍 54 🔁 21 💬 2 📌 1
Thank you! Yes, we're planning on continuing to process new data on SRA and update the resource
23.01.2025 16:04 — 👍 1 🔁 0 💬 0 📌 0
Thanks to our many collaborators who made this possible! This has been a huge effort from our lab and others, and we couldn’t have done it without all who helped.
22.01.2025 17:31 — 👍 4 🔁 0 💬 1 📌 0
Altogether, this dataset shows us how microbiomes differ throughout the world, and emphasizes the importance of more even sampling moving forward. We’re excited to continue learning from the compendium, and we plan to continue to process more samples to keep it up to date with the available data.
22.01.2025 17:31 — 👍 4 🔁 1 💬 1 📌 0
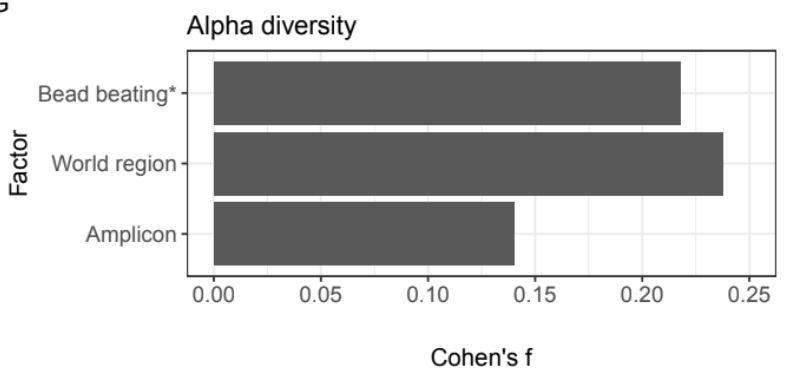
The method of DNA extraction also has a strong effect on microbiome composition. But we found that a sample’s geographic location has a bigger effect on its alpha diversity than amplicon choice or the use of bead beating, a common technique in DNA extraction.
22.01.2025 17:31 — 👍 4 🔁 0 💬 1 📌 0
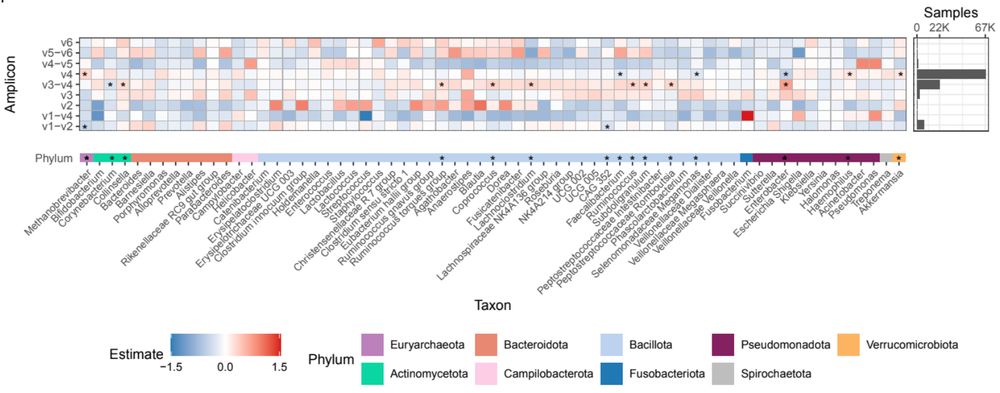
In addition to biological differences between samples, there are technical factors that can affect microbiome composition. Does the region of the 16S gene that was sequenced affect our observed results? Yes! We found genera that were differentially abundant between amplicons.
22.01.2025 17:31 — 👍 3 🔁 0 💬 1 📌 0
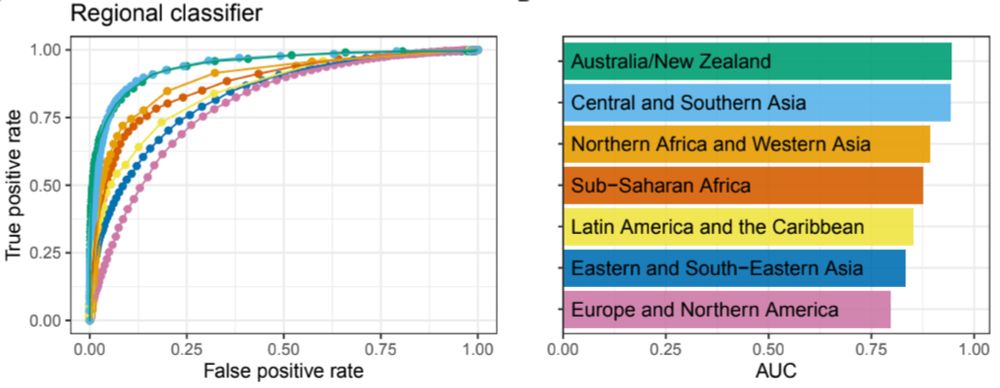
And because microbiomes from these regions are so different, we were able to train one-vs-all random forest classifiers to successfully predict where a microbiome sample came from.
22.01.2025 17:31 — 👍 1 🔁 0 💬 1 📌 0
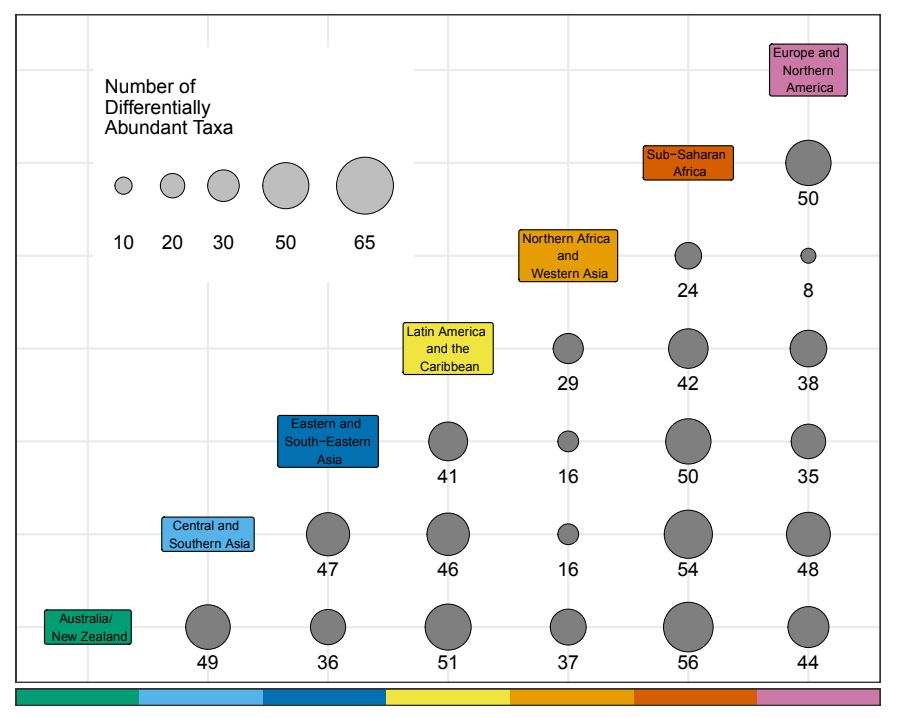
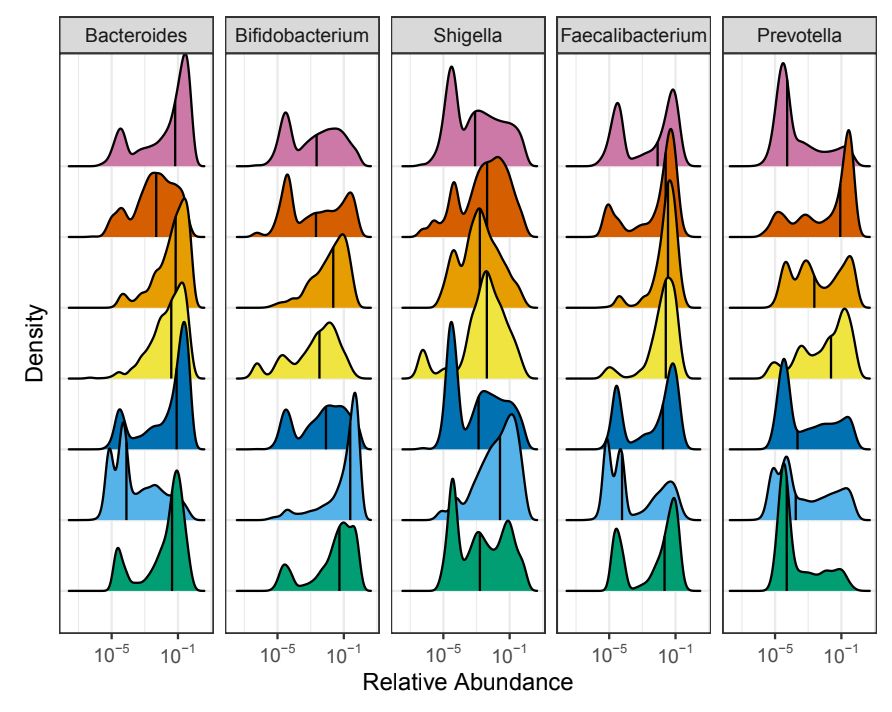
And specifically, we wanted to know what microbes were driving these large differences between world regions. We found that dozens of genera differ in abundance! Taxa like Bacteroides and Prevotella differ the most, but we found that every genus tested differed between at least two regions.
22.01.2025 17:31 — 👍 4 🔁 0 💬 1 📌 0
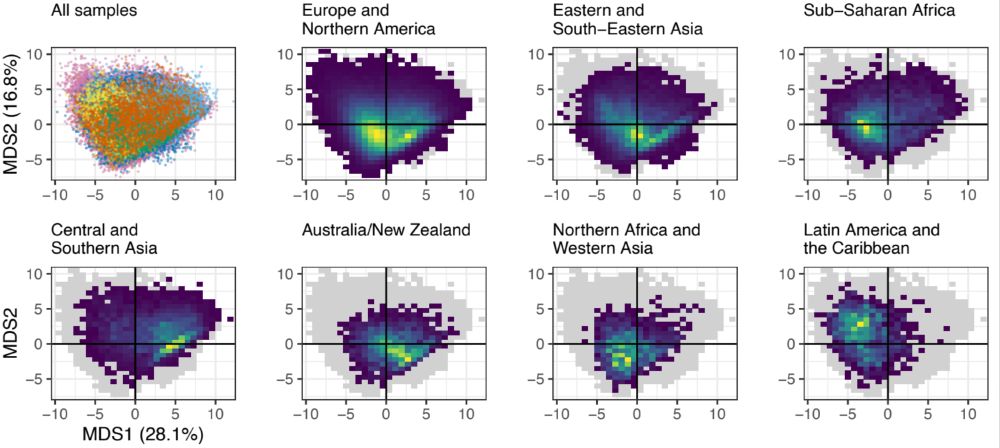
With sampling so unbalanced, are we missing a lot of information? To answer this, we wanted to see how much microbiomes differ between world regions. This big PCoA plot showed us that microbiomes from around the world have pretty drastic differences in composition.
22.01.2025 17:31 — 👍 4 🔁 0 💬 1 📌 0
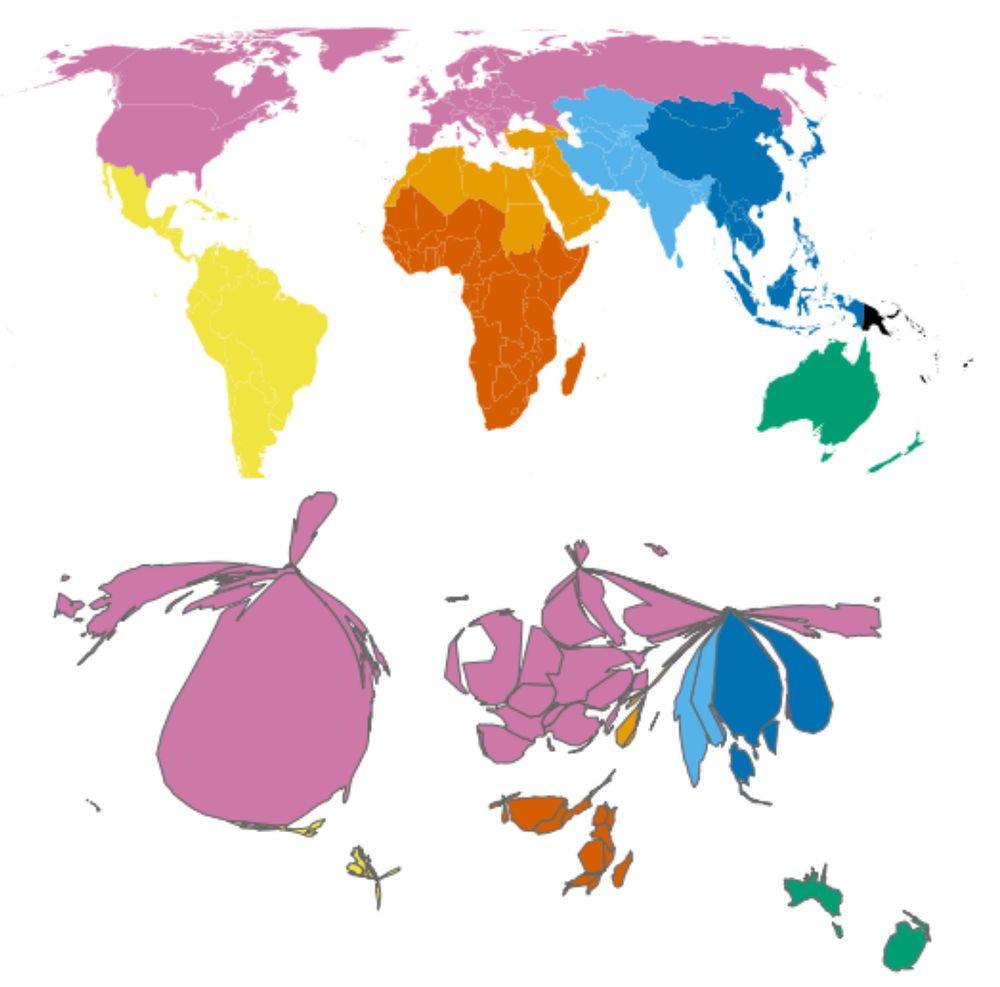
We previously found that microbiome sampling is unevenly distributed throughout the world (journals.plos.org/plosbiology/...), and we wanted to quantify just how uneven it is. Over half of our samples come from Europe/N. America, seen in this map that’s distorted to show sampling imbalance.
22.01.2025 17:31 — 👍 4 🔁 1 💬 1 📌 0
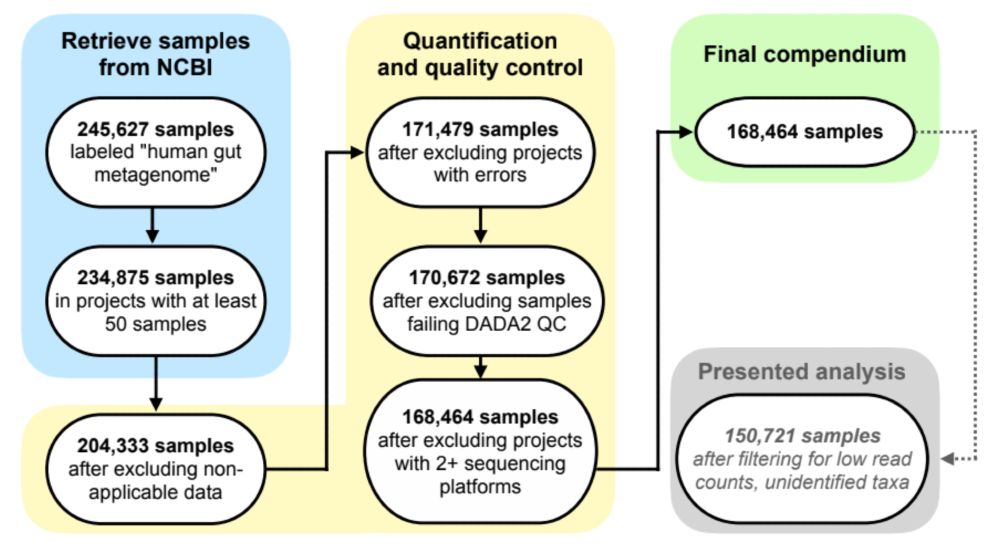
There’s tons of publicly available microbiome data, but this comes from 100s of studies that were processed separately, so we can’t directly compare results. We wanted to leverage this wealth of data, so we reprocessed raw data from 482 studies–that’s 5.7 terabases of data from all over the world!
22.01.2025 17:31 — 👍 6 🔁 0 💬 1 📌 0


attempting to start posting on here as I (hopefully) wrap up my PhD in the next several months, both about science and my one true passion--taking pictures of peanut butter sandwiches on hikes
06.12.2024 01:55 — 👍 2 🔁 0 💬 0 📌 0
PhD candidate in the Zaidi Lab at UMN | Studying mitochondrial genetics 🧬
assistant prof at University of Oregon. interested in pop gen, stat gen, human complex traits. also ELSI, metascience, ethics education, etc...
she/they. 🌈
roshnipatel.github.io
microbiome researcher | associate professor @isbscience.org | affiliate prof @uwbioe.bsky.social | ecology, evolution, and systems biology of 💩 🦠 | precision/personalized nutrition/medicine ⚕️ | 🏳️🌈 he/him | lab website: gibbons.isbscience.org
organismal biologist & geneticist | PI at med school in Houston | amazed by microbial superpowers | he/him
#microbiome #Celegans #spacebiology #firstgen 🧫 🦠 🧪 🧬
GS: https://bit.ly/goog-schol-buck-sam
ORCID: 0000-0002-4347-3997
https://mrvollger.github.io
National lab scientist studying cancer drug resistance through machine learning techniques centered around integration of mass spec (e.g. #proteomics technologies). Recreational skier/climber exploring the Pacific Northwest. Views my own.⛷️🧗🏼👩🏼🔬
Geneticist. Parent. Studying sex chromosomes and sex as a biological variable in disease.
Posts and opinions are my own.
Genetics, stats, complex traits. Asst. prof. at the University of Minnesota - Twin Cities. views my own. he/him - Zaidilab.org
PhD student in the Zaidi Lab @ University of Minnesota studying statistical and population genetics
researcher at CBMR (Copenhagen). Genomics, genetics, compbio, biostats, pun lover.
Postdoc in the Zaidi Lab @University of Minnesota (Twin Cities) | PhD @UChicago Human Genetics (Raghavan Lab) | Human pop gen, complex traits & aDNA 🧬 | 🇮🇳🇺🇸
Anthropological geneticist 🧬 Boricua 🇵🇷✊🏽Assistant Prof at University of Minnesota 🏛️Opinions are my own 🧠 I like 📚🍷🐕🦺👩🏻🔬🎶
Chicago DFI Fellow at UChicago | host-microbiome interactions, machine learning | sambhawapriya.netlify.app
CSO at Innovate Phytoceuticals. I science sometimes. Interested in all things #microbiome, #bioinformatics & data analysis.
Space tech investor at Beyond Earth Tech; mathematician, data science, tech, space, bioengineering, soccer playing and occasionally owning: The Town FC, Venezia, WPSL PRO
https://deeptechthoughts.substack.com/
https://www.linkedin.com/in/patrickleebeatty
Postdoc at the Knight Lab, UCSD. Interested in large-scale microbiome analyses at global scales.Founder of the Gut Microbiome Tree of Life Project - poopomics.com
Interested in drug development, microbial ecology, systems biology, & urbanism. I develop bacterial consortia as drugs: https://www.vedantabio.com
Research Programme Manager for Cancer Research UK. Building diverse and impactful discovery science communities.
Biochemist of 20y. Big fan of trees and wild spaces. Serial hobbyist: Theatre Tech / Climber / Caver / Softballer. ProEU. Opinions mine.
Mother & wife | Chemist (PhD @hhu.de) | Science editor & writer | DAAD @daadworldwide.bsky.social & Lindau alumna | 🐶 lover | Views all mine
https://www.linkedin.com/in/kirawelter