Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
Come join EEB at the University of Tennessee at Knoxville.
Evolutionary Genomics, Assistant Professor, Ecology & Evolutionary Biology, Fall 2026
apply.interfolio.com/170735
functional and evolutionary genomics in any system 🌱, 🦠, or 🦌 (including 👫)
28.07.2025 17:49 — 👍 3 🔁 6 💬 0 📌 0

I am Grape!
14.05.2025 19:55 — 👍 0 🔁 0 💬 0 📌 0

#MemPanG25
@thinks.lol
12.05.2025 22:08 — 👍 1 🔁 0 💬 0 📌 0


 05.05.2025 01:06 — 👍 0 🔁 0 💬 0 📌 0
05.05.2025 01:06 — 👍 0 🔁 0 💬 0 📌 0

Pangenomic context reveals the extent of intraspecific plant NLR evolution
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
6/6 Are you intrigued? Here is our work by Luisa, @kdm9.bsky.social , Max, @aconga.bsky.social
, Theresa, Leon, Justina, Christa, Oliver, Joffrey, Regina, Rosanne, @hajkdrost.bsky.social and @plantevolution.bsky.social www.biorxiv.org/content/10.1...
02.09.2024 22:17 — 👍 1 🔁 0 💬 0 📌 0
5/6 Neighborhoods "expand" & "contract" in accessions and offer a richer picture of NLR evolution in (genomic) space and (evolutionary) time. Why does it matter? Because, NLRs didn't just fell out of a 🌴, they exist in the context of all in which they live and what came before them
02.09.2024 22:14 — 👍 0 🔁 0 💬 1 📌 0
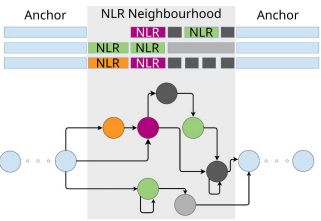
4/6 We constructed a pangenome graph with #pggb
@erikg.bsky.social to crawl along the regions of the genomes where NLRs or pieces of NLRs are found at least in 1 accession UNTIL we found common (syntenic) anchors bordering those regions in all the accessions --> "NLR neighborhood"
02.09.2024 22:13 — 👍 0 🔁 0 💬 1 📌 0
3/6 With a rigorous annotation that involved manual curation and semi-automated decision-making algorithm we pinned down every single NLR (genes, pseudogenes, "protopseudogenes") and even remnants of NLRs.
02.09.2024 22:11 — 👍 0 🔁 0 💬 1 📌 0
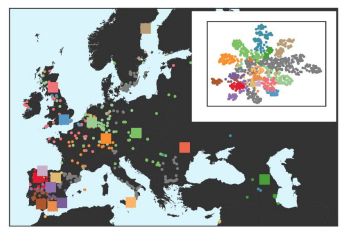
2/6 We assembled 17 diverse *A. thaliana* (Ath) genomes using @pacbio.bsky.social #CCS and annotated them using
#isoseq reads that were obtained by challenging these accessions with three different Hyaloperonospora arabidopsidis (Har) strains.
02.09.2024 22:10 — 👍 1 🔁 0 💬 1 📌 0
Ecology and Biocomplexity
On the academic job market | How are species compared to one another across different genomic regions? Postdoc at Langmead Lab, Johns Hopkins | Comparative #genomics | Phylogeny and indexing at scale | Formerly at UNIL/SIB/WUR | sinamajidian.github.io
We support #MaxPlanck researchers in founding science-based #startups by transforming cutting-edge #research into real-world solutions and incubating #innovation together.
➔ 𝗔𝗣𝗣𝗟𝗬 𝗡𝗢𝗪: http://www.maximize-incubator.com
Associate Professor @UTK_EEB & Director @UTKHerbarium | Plant development, physiology, systematics, & evolution | Member @UCWCWA | Opinions are my own
NOT SPAM. But I was reported and nothing I can do. So this account will be inactive: find me on LinkedIn, the only platform working well.
At least the bots can flourish here. Sigh.
Director General/
Natural Resources Canada
Postdoc in WeigelWorld @PlantEvolution, @MPI_bio. Evolution of Plant-Microbe-Microbe interactions, molecular evolution, and comparative genomics
The scientific editors of Cell Host & Microbe, a Cell Press journal, bring you the latest information and insights from the forefront of host-microbe research.
andreaguarracino.github.io
Assistant Professor at the University of Tennessee (@uthsc).
Ph.D. in Bioinformatics.
Computer engineer working on pangenomics.
Dad, bioinformatician, ex-expat. Currently working on transposon bioinformatics & infectious disease genomics.
Ph.D. Candidate in Plant Pathology at North Dakota State University. Member of Dr. Tim Friesen's lab at the USDA-ARS, my projects use a range of molecular biology techniques to characterize plant-fungal interaction.
Departmental Group Leader - Protein Bioinformatics @ Max Planck Institute for Biology - Tübingen @mpi-bio-fml.bsky.social
Protein evolution // MPI Bioinformatics Toolkit // prokaryotic cell-surface proteins // prokaryotic histones
Assistant Professor at University of Michigan. Evolutionary biology, spatial population genetics, dad, he/him
Chemical Biology - Plant hormone signaling - Drug Discovery - Targeted Protein Degradation
EMBO Postdoctoral Fellow || natural selection, gene expression, perennial plant-pathogen coevolution || Born at 355ppm CO2 || @mvlkova@ecoevo.social || she/her
Postdoc at ETH Zürich (Bomblies lab), PhD from Max Planck Institute for Biology Tübingen (Weigel lab). Interested in epigenomics, natural variation and the evolution of polyploidy in plants.
Postdoctoral researcher at UCL. Ancient Genomics and Evolution. https://github.com/smlatorreo
Prof of Plant Cell Biology at UC Riverside. Views are my own. She/her
Our mission is to enable the promise of genomics to better human health by creating the world’s most advanced sequencing technologies.
Genomic Data Scientist at Corteva Agriscience
BS at University of Sao Paulo, MS at Oregon State University , PhD at the University of Illinois; Views are my own.
Brazilian living in America playing with genomic data







 05.05.2025 01:06 — 👍 0 🔁 0 💬 0 📌 0
05.05.2025 01:06 — 👍 0 🔁 0 💬 0 📌 0


















