Also check out the cell fate engineering applications:
bsky.app/profile/josc...
@dominik1klein.bsky.social
ELLIS PhD student @HelmholtzMunich, Student Researcher @Apple. Interested in ML, Single-Cell Genomics, and People.
@dominik1klein.bsky.social
ELLIS PhD student @HelmholtzMunich, Student Researcher @Apple. Interested in ML, Single-Cell Genomics, and People.
Also check out the cell fate engineering applications:
bsky.app/profile/josc...
@chatgtp.bsky.social, Manuel, A. Regev, B. Treutlein, @graycamplab.bsky.social, @fabiantheis.bsky.social
23.04.2025 09:26 — 👍 1 🔁 0 💬 1 📌 0CellFlow was a highly collaborative team effort. Thanks to the great co-lead @josch1.bsky.social , and the fantastic team Daniil, Lea, Soeren, Alessandro, @le-and-er.bsky.social, Alejandro, @guillaumehu.bsky.social, @hsiuchuanlin.bsky.social, @nazbukina.bsky.social, Fatima, Theo,
23.04.2025 09:26 — 👍 2 🔁 0 💬 1 📌 0Check out the paper for more applications, including cell fate engineering and organoid protocol optimisation!
Head to cellflow.readthedocs.io for tutorials, and get in touch—Plenty of exciting directions to explore!
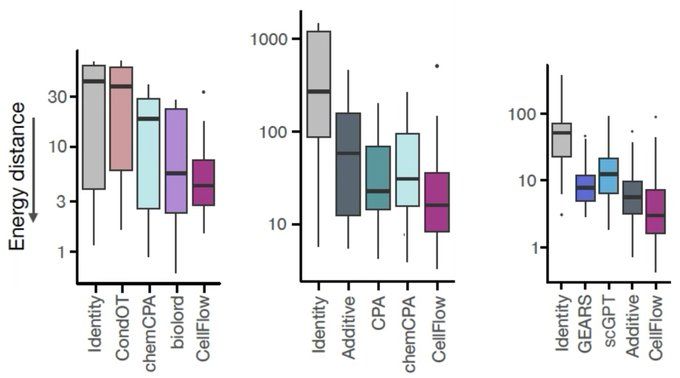
We found CellFlow to consistently perform competitively across various benchmarks, including drug and genetic perturbation screens - while being able to address complex experimental setups other methods are not able to address.
23.04.2025 09:26 — 👍 1 🔁 0 💬 1 📌 0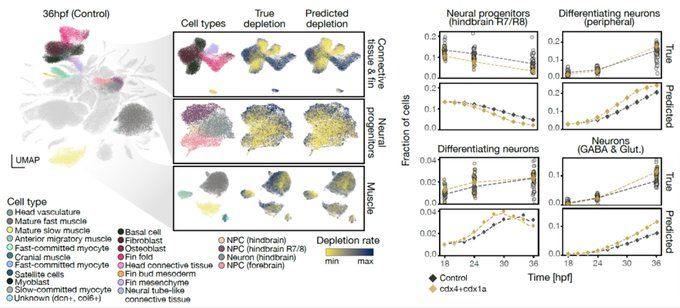
We let CellFlow learn the perturbed development of entire embryos, allowing to model the continuous trajectories of single cells under different genetic perturbations.
23.04.2025 09:26 — 👍 1 🔁 0 💬 1 📌 0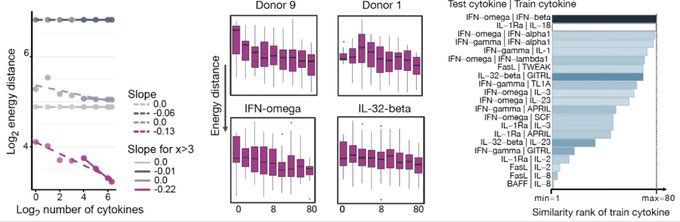
We predicted donor-specific cytokine responses on 10 million cells! We found CellFlow to exhibit scaling laws in the number of seen conditions and gained interpretable insights into model training.
23.04.2025 09:26 — 👍 1 🔁 0 💬 1 📌 0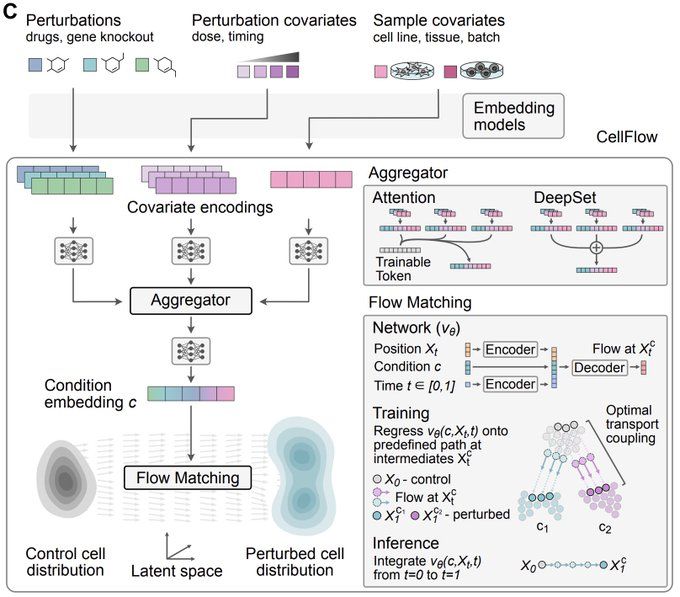
CellFlow builds on flow matching, optimal transport & attention mechanisms to learn an embedding of complex experimental conditions. This guides the flow from control to perturbed cells, generating realistic states while minimizing displacement costs.
23.04.2025 09:26 — 👍 1 🔁 0 💬 1 📌 0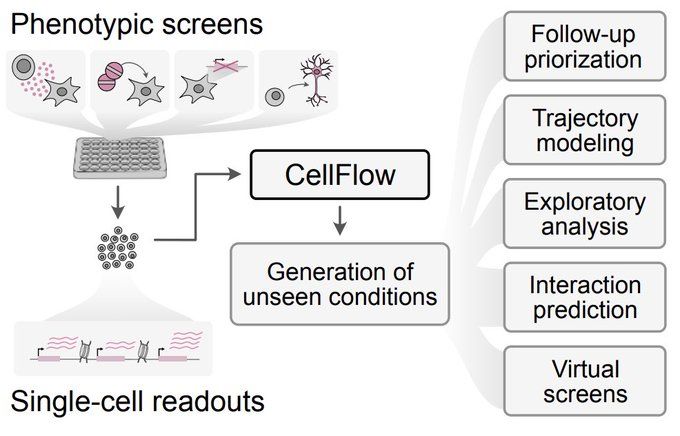
From cell lines to full embryos, drug treatments to genetic perturbations, neuron engineering to virtual organoid screens — odds are there’s something in it for you!
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1

Yay, we built a thing! With @dominik1klein.bsky.social Daniil, Aviv Regev, Barbara Treutlein @graycamplab.bsky.social @fabiantheis.bsky.social we use flow matching to enable generalised sc phenotype modeling. From cytokine screens to fate programming and organoid engineering tinyurl.com/3xhju7db
18.04.2025 10:19 — 👍 35 🔁 9 💬 1 📌 0
Excited to share our latest preprint, presenting a multi-omic human neural organoid cell atlas of the posterior brain! 🧠🔬
doi.org/10.1101/2025...
Great work with @hsiuchuanlin.bsky.social @zhisonghe.bsky.social @graycamplab.bsky.social and Barbara Treutlein!
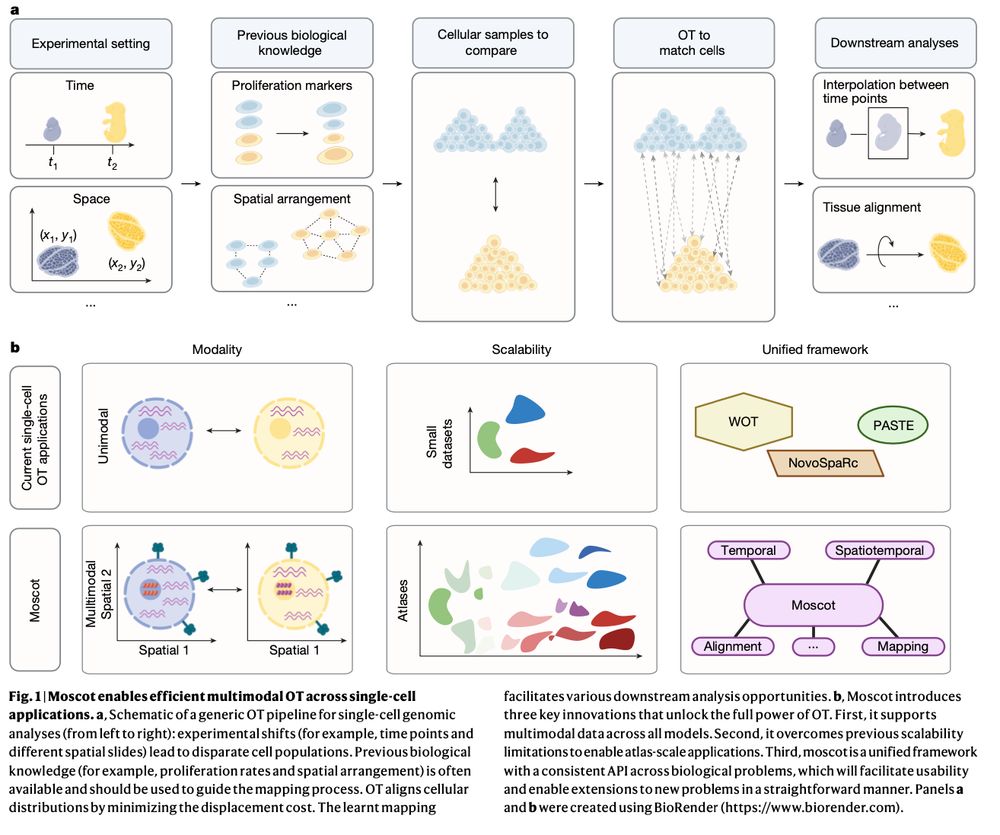
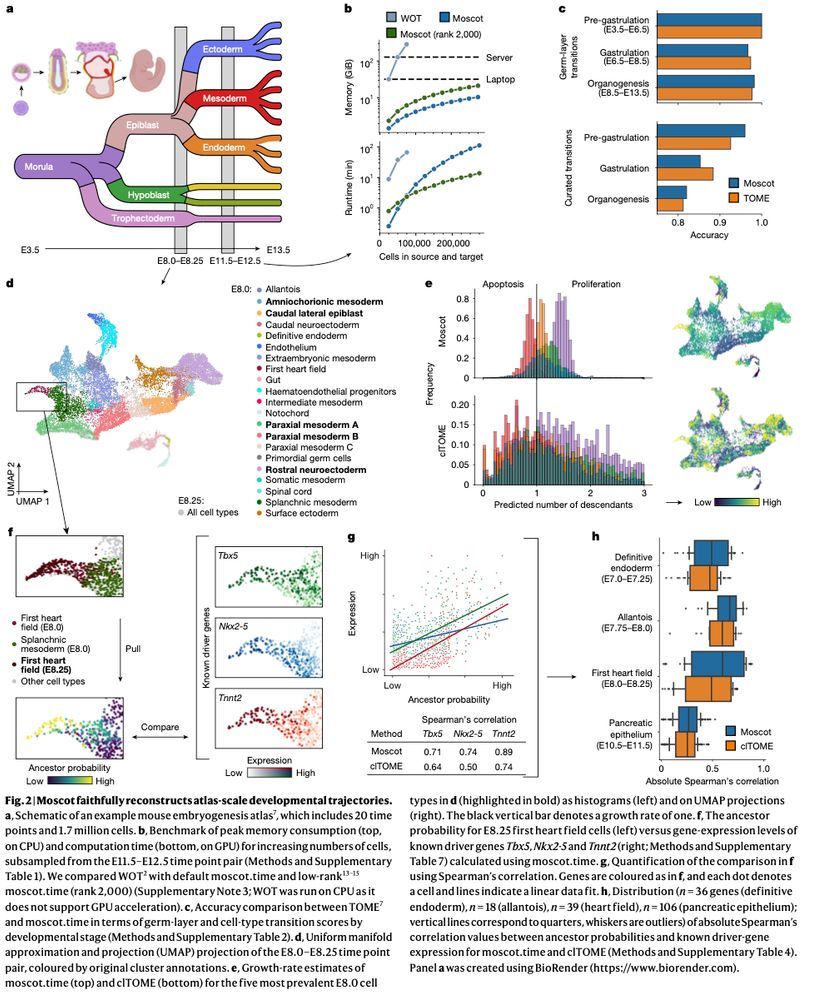
Mapping cells through time and space with moscot https://www.nature.com/articles/s41586-024-08453-2 🧬🖥️🧪 https://github.com/theislab/moscot
23.01.2025 21:00 — 👍 13 🔁 5 💬 0 📌 0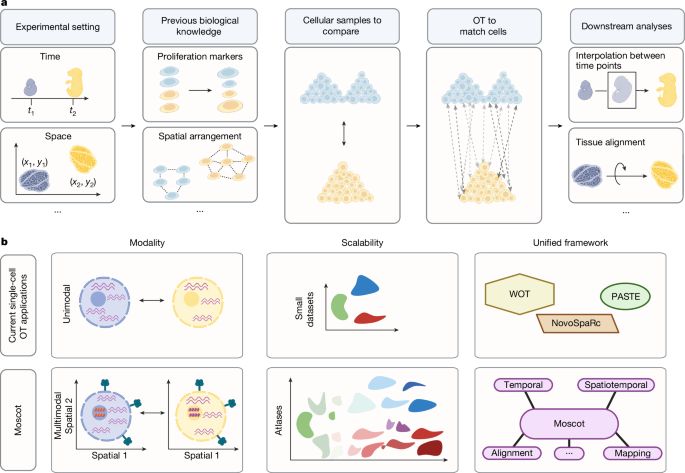
Nature research paper: Mapping cells through time and space with moscot
https://go.nature.com/40pqpvw
Missing the deep learning part? go check out the follow up work @neuripsconf.bsky.social (tinyurl.com/yvf72kzf) and @iclr-conf.bsky.social (tinyurl.com/4vh8vuzk)
23.01.2025 08:45 — 👍 11 🔁 3 💬 0 📌 0@AimeeBastidas, @pacotael, @MartaTarquis, @ShreyParikh07, Ilan Gold, @heikolickert.bsky.social , @mostafabakhti.bsky.social @marcocuturi.bsky.social , @fabiantheis.bsky.social
23.01.2025 08:41 — 👍 4 🔁 1 💬 0 📌 0This was a highly collaborative project, thanks to everyone involved, in particular to the co-leads @giopll.bsky.social , @mariuslange.bsky.social , Michal Klein, Zoe Piran, as well as Manuel Gander, @Laetitia_Ppx, Michael Sterr, @lamasa LamaSaber95, @Diana61204366,
23.01.2025 08:41 — 👍 4 🔁 0 💬 1 📌 0moscot integrates straightforwardly with the @scverse.bsky.social ecosystem. Check out our tutorials and examples at moscot-tools.org to analyze your single-cell data, we encourage the community to contribute to moscot with novel OT applications!
23.01.2025 08:41 — 👍 4 🔁 1 💬 1 📌 0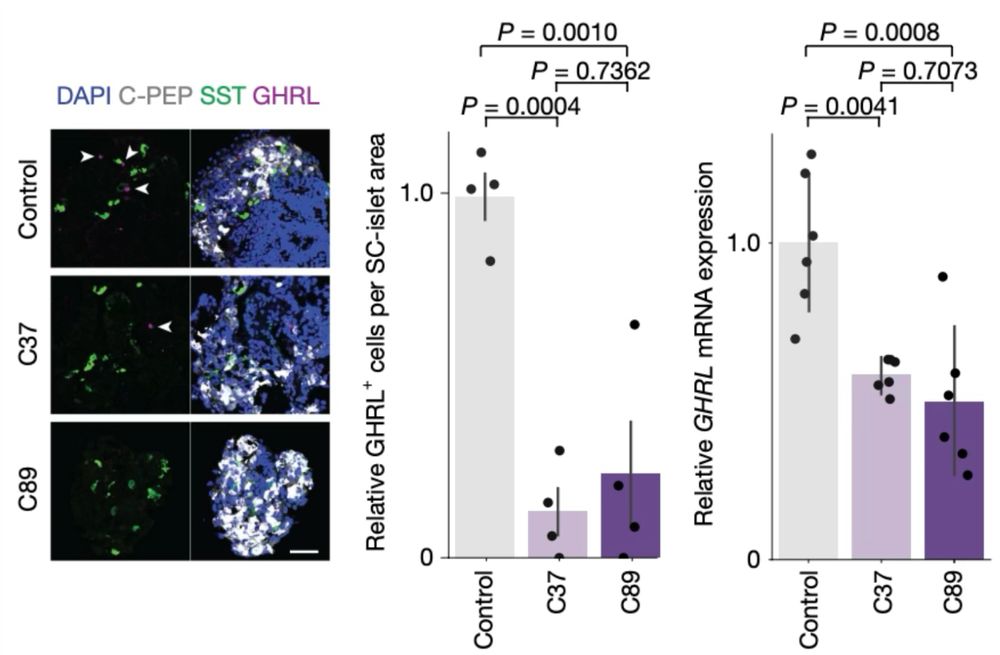
moscot’s ability to study gene regulation allowed us to hypothesise Neurod2 to be an activator of epsilon cell formation. We indeed observed a reduction in ghrelin (hormone produced by eps. cells) in NEUROD2 knockout iPSCs
23.01.2025 08:41 — 👍 4 🔁 0 💬 1 📌 0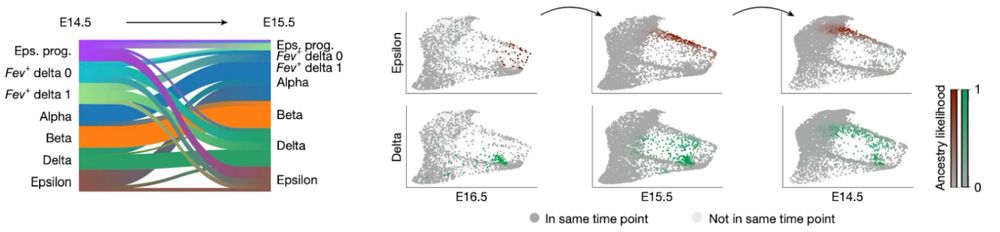
We generated a new dataset of the developing mouse pancreas across three time points with paired measurements of gene expression and ATAC data. Enrichment of Ngn3 high cells allows us to disentangle the poorly understood formation of delta and epsilon cells.
23.01.2025 08:41 — 👍 3 🔁 0 💬 1 📌 0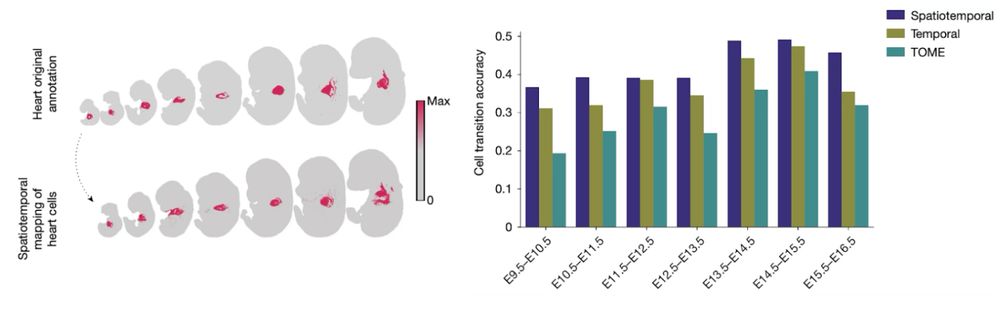
We develop a new method for the analysis of spatiotemporal datasets. We show how incorporating the spatial information improves the recovery of cell trajectories and demonstrate its use in a spatially resolved mouse embryogenesis dataset.
23.01.2025 08:41 — 👍 3 🔁 0 💬 1 📌 0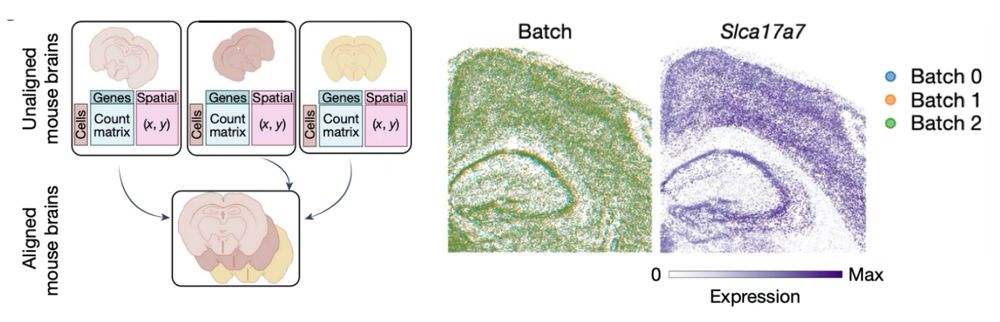
Moscot aligns large-scale spatial transcriptomics slides from different individuals to obtain a statistically more profound representation of the mouse brain.
23.01.2025 08:41 — 👍 3 🔁 0 💬 1 📌 0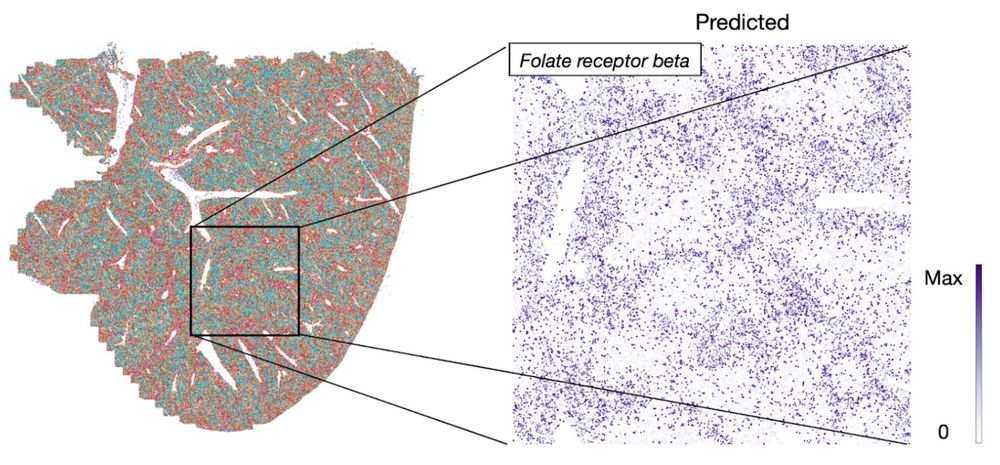
We map CITE-seq data of the mouse liver to a spatial reference slide incorporating information from gene expression, protein, and spatial measurements, facilitating liver zonation.
23.01.2025 08:41 — 👍 3 🔁 0 💬 1 📌 0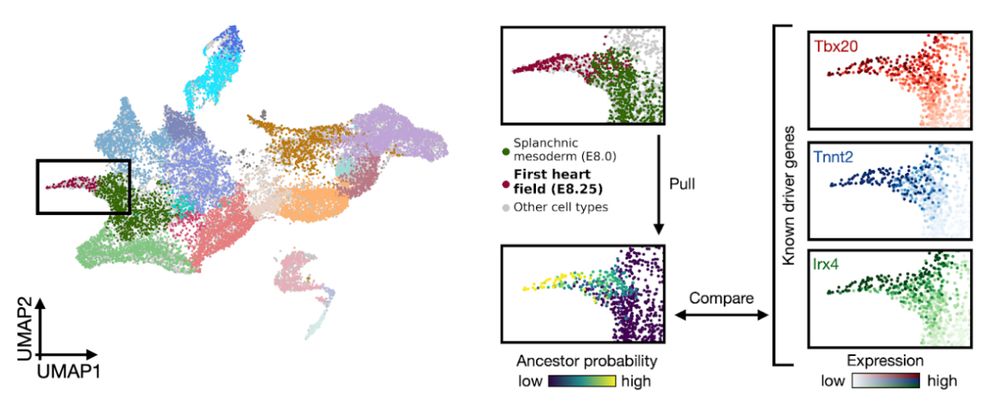
Incorporating recent advances in OT, we make moscot scalable to atlas-scale datasets (see ott-jax.readthedocs.io/en/latest/, @marcocuturi.bsky.social). This allows to to recover trajectories in an atlas of mouse embryogenesis comprising 1.7 million cells.
23.01.2025 08:41 — 👍 3 🔁 0 💬 1 📌 0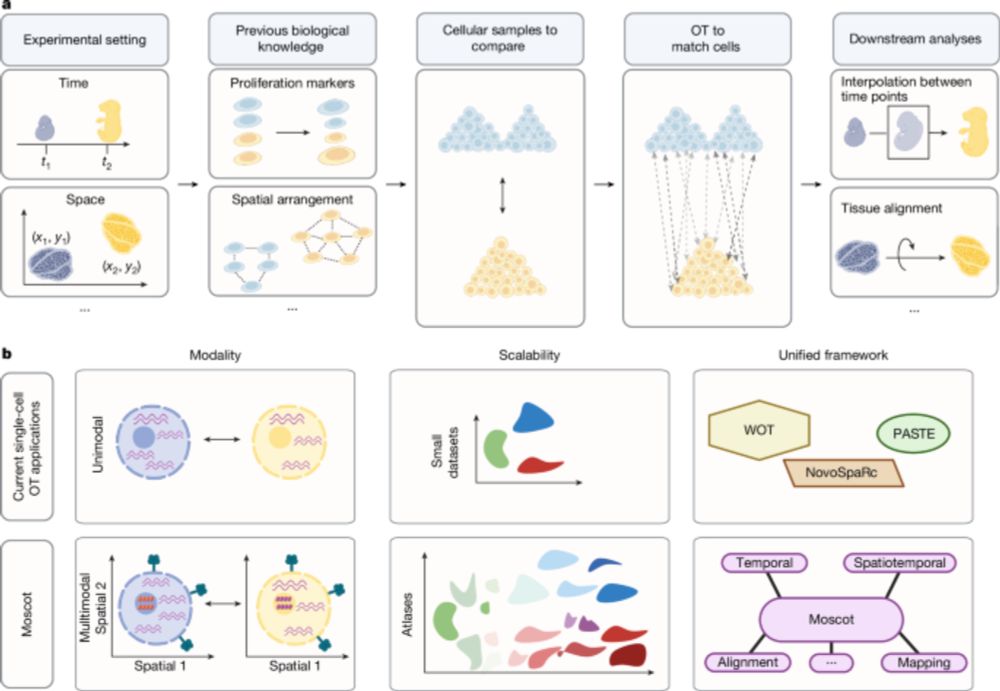
Good to see moscot-tools.org published in @nature.com ! We made existing Optimal Transport (OT) applications in single-cell genomics scalable and multimodal, added a novel spatiotemporal trajectory inference method and found exciting new biology in the pancreas! tinyurl.com/33zuwsep
23.01.2025 08:41 — 👍 49 🔁 13 💬 1 📌 3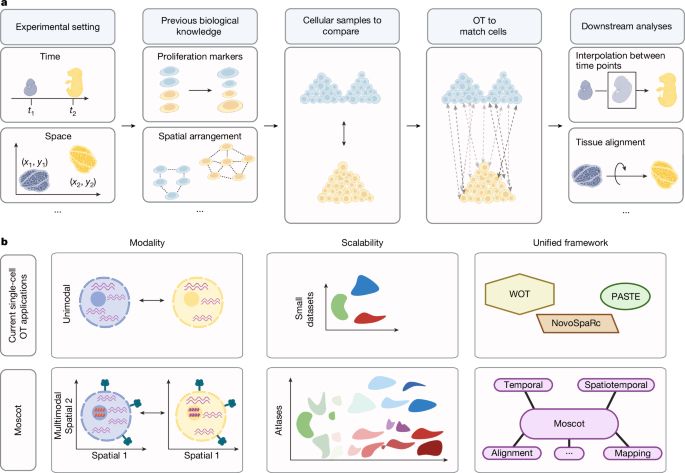
Excited to see Moscot (moscot-tools.org) published in @Nature! We scaled Optimal Transport (OT) in single-cell genomics & added multimodality together with spatiotemporal trajectory inference, finding exciting new biology in the pancreas! 🚀 Read at www.nature.com/articles/s41...
22.01.2025 22:46 — 👍 126 🔁 42 💬 2 📌 2Exciting new tool to make the most out of multimodal single-cell data from our friends and long-term collaborators of the @fabian_theis lab - check it out, it works beautifully and really makes sense out of your complex data! 👇👍😉
23.01.2025 06:43 — 👍 26 🔁 10 💬 0 📌 0Today is a great day for optimal transport 🎉! Lots of gratitude 🙏 for all folks who contributed to ott-jax.readthedocs.io and pushed for the MOSCOT (now @ nature!) paper, from visionaries @dominik1klein.bsky.social, G. Palla, Z. Piran to the magician, Michal Klein! ❤️
www.nature.com/articles/s41...
1/2 🚀 Discrete Optimal Transport (OT) has proved useful in a wide variety of tasks in single-cell genomics, see e.g. moscot-tools.org. Can we combine this powerful concept this with novel generative models? 🤔
🌟 Enter GENOT: Generative Entropic Neural OT.