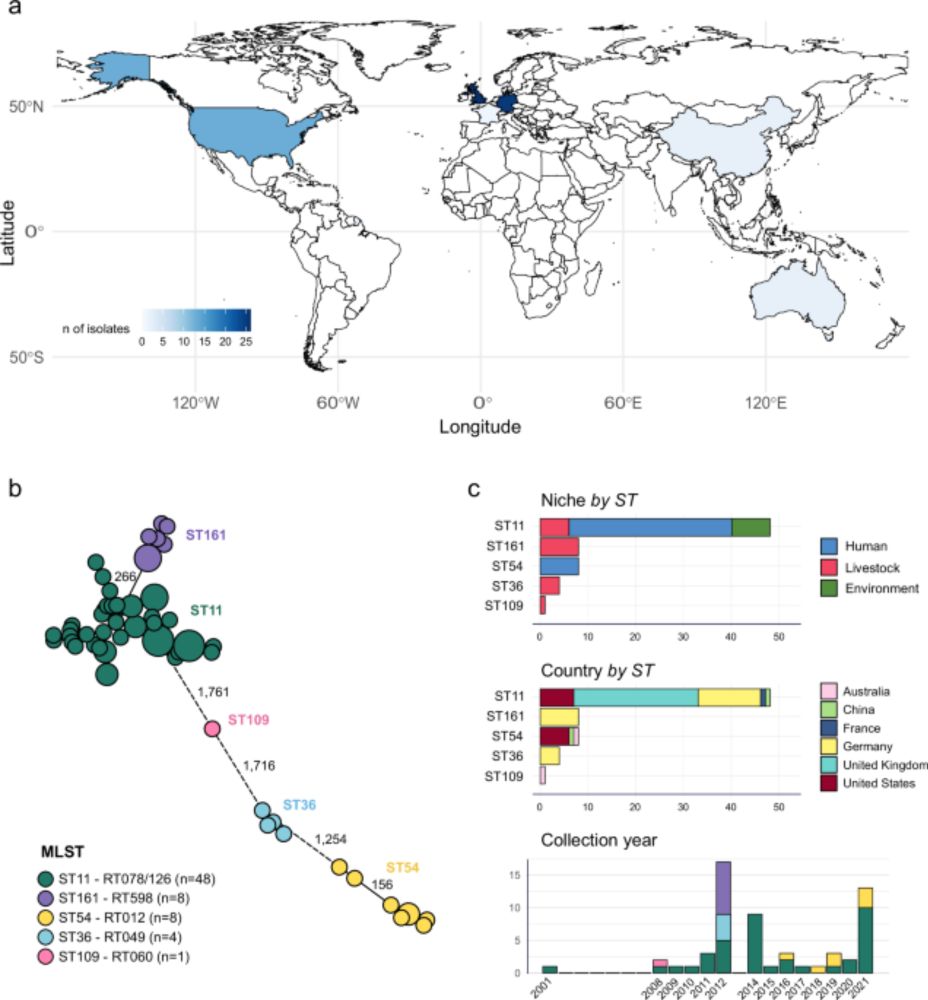
Super cool story from Sam Fenn. He developed a wound model where s. aureus & p. aerunginosa & c. albicans fight it out shows that copper is central to the outcome where Candida and pseudomonas take copper from human tissue to gang up on staph!
But staph being staph is as always one step ahead….
11.02.2026 08:41 — 👍 13 🔁 5 💬 2 📌 1

Bioinformatician Position Open at the Evolutionary and Functional Genomics Lab @ibb-botanic.bsky.social and Nuria Flames' group @ibv-csic.bsky.social
If you are interested, please send a CV and a short motivation letter to nflames@ibv.csic.es with the subject “Bioinfo position 2026” before 27/02/26
28.01.2026 14:00 — 👍 2 🔁 2 💬 0 📌 0

My team at @cbitoulouse.bsky.social is recruiting a postdoc #bioinformatics with solid experience in metagenomic analyses.
Interest in evolution, ecology & MGEs is important.
The offer stands until the perfect candidate is found, and it could be you 🫵
🔁 🙏
#microSky #phagesky #UTIsky
@cnrs.fr
15.01.2026 11:19 — 👍 27 🔁 55 💬 1 📌 0
Bacteria chromosomes contain Genomic Islands that provide virulence, antibiotic resistance, MGE-defence,... They transfer between cells, but the mechanism of most remains elusive.
Here we explore the conjugative capacity of these mysterious Genomic Islands.
www.biorxiv.org/content/10.6...
14.01.2026 10:14 — 👍 82 🔁 52 💬 4 📌 2
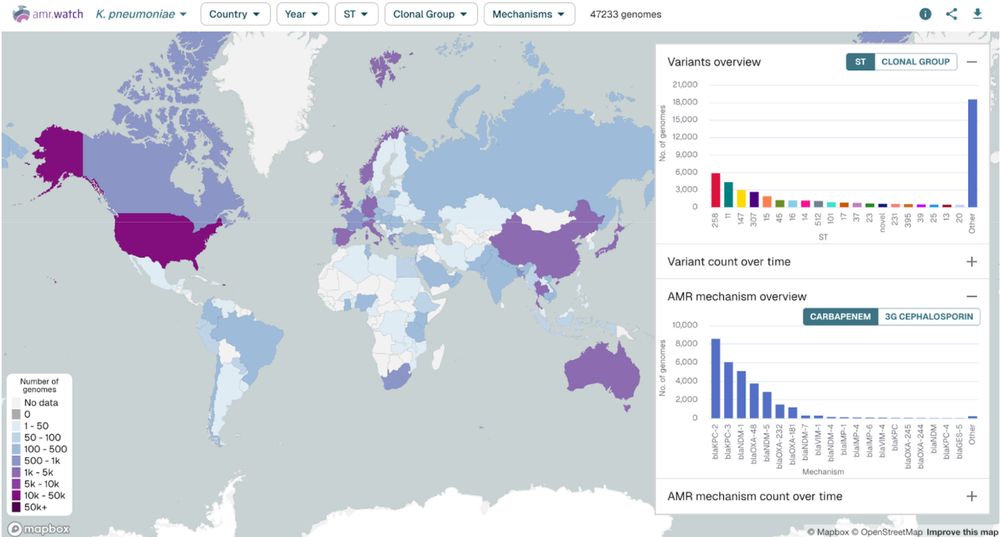
Our latest preprint is out describing amr.watch - a platform for monitoring AMR trends from global genomics data: www.biorxiv.org/content/10.1... (1/6)
22.04.2025 09:54 — 👍 34 🔁 23 💬 1 📌 5

Foto de grupo

Mesa 1

Mesa 2

Programa
Hoy hemos celebrado la Asamblea Anual de la Conexión Resistencias a Antimicrobianos del @csic.es donde nos hemos reunido para hablar sobre este problema de salud y buscar soluciones. Una conexión liderada por @albertomarina.bsky.social y Ana de la Torre. Síguenos para estar informado!
24.11.2025 18:48 — 👍 14 🔁 6 💬 1 📌 1
Happy to share our new AMR resource which has phenotypic AMR (usually MIC data) collected from publications and databases. This is paired with assemblies and annotations
We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use
19.11.2025 12:27 — 👍 62 🔁 36 💬 1 📌 0
2026-27 Project (Dyson & Mostowy & Knight) - MRC London Intercollegiate Doctoral Training Partnership Studentships
PHACTS: Unravelling PHAge-baCTeria-host immune dynamicS to inform phage therapy SUPERVISORY TEAM Supervisor Dr Zoe Dyson...
London interdisciplinary #PhD position now open with myself, @sergemostowylab.bsky.social, & @gmknght.bsky.social on #phage -bacteria-host immune dynamics for WHO priority bacterial pathogens #Klebsiella, #Shigella, and #Staph. Combines cellular microbiology, genomics, & mathematical modelling.
13.11.2025 11:11 — 👍 9 🔁 12 💬 0 📌 0

We wrapped up #IMMEM XIV over the weekend and it was a fantastic success with over 250 participants from 35 countries. A big thank you to the organisers and specifically @esgem-sg.bsky.social our study group on epidemiological markers! The content will be available on ESCMID media soon. #clinmicro
22.09.2025 06:54 — 👍 14 🔁 5 💬 0 📌 0

Efficient sequence alignment against millions of prokaryotic genomes with LexicMap - Nature Biotechnology
LexicMap uses a fixed set of probes to efficiently query gene sequences for fast and low-memory alignment.
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
10.09.2025 09:12 — 👍 190 🔁 99 💬 5 📌 4
Our high-precision metagenomic strain caller, PHLAME, is now published in Cell Reports!! www.cell.com/cell-reports...
PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
15.08.2025 16:33 — 👍 47 🔁 26 💬 2 📌 0
Very exciting work here from Sanjana Kulkarni, Maha Farhat and team. CNN prediction of MIC, v thoroughwork!!
14.08.2025 07:26 — 👍 4 🔁 4 💬 0 📌 0

Register today for the 14th International Meeting on Microbial Epidemiological Markers (IMMEM XIV)! Topics range from how genomics inform resistance profiling to pathogen surveillance in a #OneHealth framework. 🧬
✍️ Register: ow.ly/gjOZ50WpOXX
👀 Programme: ow.ly/Whj850WpOXY
#IDSky
07.08.2025 08:14 — 👍 5 🔁 6 💬 0 📌 1

Researchers at @lshtm.bsky.social, led by @josebengoechea.bsky.social, will conduct a £2.2M study funded by UK MRC to investigate how Klebsiella pneumoniae silently colonises the gut before causing deadly infections. 🔬🦠
Read more 👉 bit.ly/4mNSreh
05.06.2025 13:17 — 👍 27 🔁 9 💬 1 📌 1

Invited & Abstract Programme
Learn more about the congress and events hosted by ESCMID in the fields of Clinical Microbiology (CM) and Infectious Disease (ID).
Late breaker abstracts for IMMEM conference, to be held in beautiful Portugal this September, close Friday May 23!
Something for the #ABPHM25 crowd to ponder 🤔
www.escmid.org/congress-eve...
21.05.2025 16:57 — 👍 4 🔁 3 💬 0 📌 0
🧬Antibiograms have been used during outbreak investigations for decades as a surrogate for genetic relatedness of #MRSA
🦠Authors evaluate the accuracy of antibiograms in detecting transmission, using genomic epidemiology as the reference standard
www.sciencedirect.com/science/arti...
20.02.2025 13:50 — 👍 2 🔁 1 💬 0 📌 0
Composite of the paper header showing all authors and an equation showing phylogenetic trees plus plasmids give a complex plasmid picture.
📣Our latest paper is out now in mSystems: Not quite as it may seem - investigating #plasmid diversity and evolution in #Klebsiella pneumoniae at a single institution over time. A short thread 🧵 @zaminiqbal.bsky.social
14.01.2025 14:20 — 👍 66 🔁 44 💬 2 📌 2
Many thanks to all co-authors contributed to this work: Beth Blane, Katie Bellis, Marta Matuszewska, Toska Wonfor, Dorota Jamrozy, Michelle Toleman, Joan Geoghegan, @julianparkhill.bsky.social, Ruth Massey, Sharon Peacock, @ewanharrison.bsky.social
14.01.2025 09:40 — 👍 1 🔁 0 💬 0 📌 0
Our study also identified several uncharacterised genes that may be critical for colonisation that warrant further experimental investigation.
14.01.2025 09:40 — 👍 0 🔁 0 💬 1 📌 0
We also found mutations in genes that could influence how S. aureus interact with human cells or the immune system, such as in genes encoding SraP/SasA and TarS.
14.01.2025 09:40 — 👍 0 🔁 0 💬 1 📌 0
Nitrogen metabolism showed the strongest evidence of adaptation, with genes encoding sub-units of the assimilatory nitrite reductase (nasD/nirB) and urease (ureG), and a nitrogen regulatory protein (darA/pstA), showing the highest mutational enrichment.
14.01.2025 09:40 — 👍 0 🔁 0 💬 1 📌 0
We observed an excess of protein-altering mutations in metabolic genes, in the Agr system and in antibiotic targets; and showed the phenotypic effect of multiple adaptive mutations including changes in haemolytic activity, antibiotic susceptibility, and metabolite utilisation.
14.01.2025 09:40 — 👍 0 🔁 0 💬 1 📌 0
We provide data, insight and tools for pathogen surveillance and AMR research. Based at the Pandemic Sciences Institute, University of Oxford, UK.
Ecology and evolution of antibiotic resistance at CNB-CSIC.
@besaraornottobe on X 🐤
La resistencia a los antimicrobianos representa una amenaza creciente para la sociedad. En AMR Hub CSIC impulsamos soluciones innovadoras para frenarla y garantizar la eficacia de los antibióticos, tanto hoy como en el futuro.
Lead Clinical Bioinformatician in Pathogen Sequencing at St Thomas' Hospital in London, UK (Synnovis).
Working on infectious disease diagnostics, pathogen genomics and metagenomics.
Assistant prof @ UofT, associated scientist @ Helmholtz Institute for RNA-based Infection Research. Pathogen systems biology / informatics / functional genomics. Coastal New Hampshirite. Drink Moxie.
Group Leader at KCL studying host pathogen interactions
Senior Researcher at Uppsala University | Host-pathogen interactions | Intestinal organoids | Virulence gene regulation | Shigella and Salmonella | Functional genomics | Affiliated to the SellinLab @sellinlab.bsky.social
Dynamics of Host-Pathogen Interactions Unit at @pasteur.fr https://research.pasteur.fr/en/team/dynamics-of-host-pathogen-interactions/
Bacterial geneticist 🧫 🦠 🧬 focusing on #Shigella, #enteric #pathogens & #phage. Using host signals & #organoids to study #pathogenesis. Assist Prof at USUHS. Views my own. https://medschool.usuhs.edu/profile/christina-faherty-phd
Group leader and Wellcome Trust Career Development Fellow at King's College London. Using comparative genomics, microbiology and innate immunology to study Klebsiella virulence. Foodie and below average footballer.
Researching the diversity and interaction of phages with plant pathogenic bacteria 🌱🔬 | INRAE | PHIM | Montpellier, France. 🇵🇸
Professeure/chaire d'excellence du Canada
Université de Montréal
Département de microbiologie, infectiologie et immunologie
What do I like? Wild things 😜 yet phages are my favorite!
The Continent is an award-winning weekly newspaper made by African journalists. Free on 👇
📱WhatsApp: bit.ly/4b2gpfI
💬 Signal: bit.ly/TCSignal
📲 Telegram: t.me/continentnews
✉️ Email: read@thecontinent.org
👍 Donate: http://bit.ly/3IYcp6X
Sequity helps labs turn their genomics goals into real, impactful outcomes.
We believe microbial genomics should be both accessible and sustainable in every context - and we work shoulder-to-shoulder with you to make that happen.
www.sequity.bio
Evolution of antibiotic resistance. Ramón y Cajal Fellow at Universidad Autónoma de Madrid. https://amrevolution.es/
Postdoctoral researcher at Intistut Pasteur | MSCActions | Vet | Plasmid Biology, Bacterial Evolution & Antimicrobial Resistance
PhD student at UCM
Molecular microbiologist
MGE and bacterial adaptation